Abstract
Introduction
While pharmaceutical companies aim to leverage real-world data (RWD) to bridge the gap between clinical drug development and real-world patient outcomes, extant research has mainly focused on the use of social media in a post-approval safety-surveillance setting. Recent regulatory and technological developments indicate that social media may serve as a rich source to expand the evidence base to pre-approval and drug development activities. However, use cases related to drug development have been largely omitted, thereby missing some of the benefits of RWD. In addition, an applied end-to-end understanding of RWD rooted in both industry and regulations is lacking.
Objective
We aimed to investigate how social media can be used as a source of RWD to support regulatory decision making and drug development in the pharmaceutical industry. We aimed to specifically explore the data pipeline and examine how social-media derived RWD can align with regulatory guidance from the US Food and Drug Administration and industry needs.
Methods
A machine learning pipeline was developed to extract patient insights related to anticoagulants from X (Twitter) data. These findings were then analysed from an industry perspective, and complemented by interviews with professionals from a pharmaceutical company.
Results
The analysis reveals several use cases where RWD derived from social media can be beneficial, particularly in generating hypotheses around patient and therapeutic area needs. We also note certain limitations of social media data, particularly around inferring causality.
Conclusions
Social media display considerable potential as a source of RWD for guiding efforts in pharmaceutical drug development and pre-approval settings. Although further regulatory guidance on the use of social media for RWD is needed to encourage its use, regulatory and technological developments are suggested to warrant at least exploratory uses for drug development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Social media displays utility for better understanding real-world patient outcomes and experiences. |
There is a gap in understanding of how social media can be used for drug development, both pre-approval (e.g. to guide clinical trial development and to observe similar competitor products) and post-approval (e.g. accounting for off-label use or developing patient education material). Extant research has primarily focused on using social media data for safety surveillance after it has been released to the market. |
Regulations accounting for this are incomplete and require updating to incentivise industry and academia. |
1 Introduction
1.1 Real-World Data (RWD) and Social Media
The evidence base for pharmaceutical products, typically randomised clinical trials, aims to ensure patient safety and drug efficacy. However, in recent years, it has come under increasing scrutiny. On the one hand, conventional randomised clinical trials have been shown to not always reflect the demographics of real-world patient populations [1]. On the other hand, observing drug usage in the real world, outside randomised clinical trials, is emerging as a potential means for more targeted ways of informing drug efficacy and safety profiles [2, 3]. Observing real-time usage is also important for drugs that are used off-label, where the evidence of the appropriateness of the drug or the dose for treating a condition is scarce [4]. As the appreciation of the gap between clinical drug development and real-world treatment outcomes becomes apparent, adopting novel means of understanding patient outcomes and perspectives becomes increasingly important for both pharmaceutical and technology companies, which are investing tens of billions of dollars in health-related data analysis capabilities [5].
Recent years have seen a wave of efforts to bridge this gap. In 2016, the USA passed the 21st Century Cures Act designed to accelerate drug development and, among other goals, modernise the collection and analysis of drug efficacy and safety data [6]. This coincided with a patient-centric paradigm shift in both drug development and safety monitoring, often centred around real-world data (RWD), i.e. the various patient data collected outside of traditional clinical development. Examples include using electronic healthcare records and insurance claims to understand patient responses to aspirin doses [3], and the use of RWD to supplement regulatory approvals of therapies for diffuse large B-cell lymphoma based on clinical trials [7]. The need for additional data sources has also been linked to under-reporting of adverse drug reactions (ADRs), with a staggering 90–94% of such reactions being unreported [8].
Unstructured and spontaneously recorded, often digitally, patient health information is distinct from RWD collected from electronic healthcare records or other forms of structured health data, and has been suggested as a possible supplementary source to traditional data captured in clinical trials [9]. One such source garnering increasing attention is social media [10]. Sparked by the availability of data and the development of advanced analytical tools, many researchers have turned to social media to collect patient health information [11,12,13]. Today, machine learning and natural language processing (NLP) algorithms can effectively find relevant information in user-generated content on social networks or in patient health forums. Prior work has, for instance, targeted social media to extract patient health information on adverse events (AEs) [14] or drug–drug interactions [15]. Subsequently, using social media for RWD emerges as a potential addition to the drug developer’s toolbox for understanding patient needs and supporting product claims.
Several studies have demonstrated the feasibility of collecting RWD from social media [16,17,18,19]. However, in the scope of RWD and RWE, most research so far has focused almost exclusively on applications for safety monitoring. Hence, social media has primarily been situated in the context of detecting safety events after a drug has been developed and marketed. This means that the potential utility of social media for RWD in the drug development setting needs more research. “Development” here refers both to pre-approval activities and to any changes performed post-approval to increase the scope of drug applicability or enhance patient understanding of the drug. The need to focus on development is also highlighted by the fact that the US Food and Drug Administration (FDA) only relatively recently recognised the use of social media to support earlier pre-approval stages of drug development [9]. As regulatory guidance is relatively new, its applications in practice are yet unclear. In addition, extant research has typically focused separately on developing the technology for mining social media, conceptualising the medium’s limitations and opportunities, or discussing the regulatory landscape for RWD. These themes are yet to be synthesised in a single piece of work.
This paper addresses these gaps by investigating how social media can be used as a source of RWD in drug development to support regulatory decision making. It develops a machine learning pipeline to identify AEs and potential patient conditions related to anticoagulants from X (previously called Twitter) data. These findings are analysed from the industry perspective of regulatory decision making, complemented by interviews with professionals from a pharmaceutical company, against the backdrop of recent regulatory guidance from the FDA. Use cases for social media RWD are proposed. It concludes on the utility of social media for regulatory decision making, implications for industry and regulators, and addresses remaining gaps from both an industry and regulatory point of view.
The contribution of this paper is two-fold. First, it proposes actionable use cases and next steps to be further explored by industry, regulators and researchers. This is achieved by investigating how insights derived from social media data can be used to observe drug effects possibly outside clinical trials in an industry setting, given recent developments in technology, industry practice and regulatory maturity. Second, it highlights key elements in the end-to-end data pipeline and, in doing so, removes some of the previously argued barriers for utilising social media, such as a low likelihood of detection of early safety signals. Of course, we do not argue for a replacement of clinical trials with data collected from social media, or for generating evidence from social media posts. Instead, social media is seen as a low-burden real-time complement to other forms of collecting RWD.
1.2 Social Media and Health Information
Social media has so far been mainly conceptualised in the scope of pharmacovigilance. Recent years have seen increasing efforts to expand the traditional evidence base for drug safety monitoring, and social media has likewise emerged as a potential source for identifying safety signals [10, 13, 20]. Although this paper pursues a different goal than just safety monitoring—focusing on the regulatory activities related to the actual development of the drug rather than safety measures once it is marketed—scrutinising past work in this area sheds light on the opportunities and challenges of social media.
The promise of social media as a source of health information is first owing to the size of data available and real-time velocity of the data. Social media offers unprecedented volumes of information that can be captured [10], of course with appropriate ethical considerations and regulatory requirements (such as General Data Protection Regulation). For instance, the majority of US Internet users have been observed to use the Internet for discussing health information and posting about specific symptoms, drug effects or general healthcare experiences [21].
In addition, social media open doors for real-time monitoring and signal detection [22]. Past research reported results of health monitoring on various social media platforms, including Facebook [19], Instagram [16], Reddit [23] and X [17]. Of those, X has been a popular platform for collecting health data. One review of 128 articles on social media-based surveillance systems that use machine learning for real-time disease prediction found that 64% of those articles used it as their data source [24]. X’s large user base, real-time nature and the increasing openness of the public towards posting about adverse drug effects contribute to its potential for immediate scalable access to monitoring drug symptoms for pharmaceutical companies and regulators [17, 20].
Social media can also aid in understanding the uses of a drug post-approval, after it has been launched. Take off-label drug use, for example. Physicians frequently prescribe drugs for uses other than what these drugs were approved for by the regulators. Moreover, this practice is legal from at least the FDA perspective [25]. Examples of drugs that have been widely used off-label include treatment for diabetes mellitus used for cosmetic weight loss [26], drugs for malaria and parasites used to treat coronavirus disease 2019 [27], and aspirin to prevent heart attacks [25]. Data collected from social media can be used to identify patterns in off-label drug use on social platforms such as Tinder [28] and Reddit [29].
Increased data availability is complemented by advances in NLP and information extraction. These have become increasingly important to the field of biomedicine, emerging as tools for researchers and clinicians to support patient safety [30]. The development of pre-trained language models such as BERT [31] that can be adapted to the domain and task of interest have made it possible to achieve state-of-the-art performance on medical datasets [32]. More recent large language models may also prove beneficial for this analysis [33]. Commercially available NLP systems including I2e or MetaMap are also capable of identifying AEs in patients’ language and translating these into medical ontologies such as MedDRA terminology [34]. It is thus now possible to effectively deal with the large amounts of social media data and make sense of its otherwise unstructured and colloquial content [35].
By now, the tools and processes for leveraging social media for pharmacovigilance or other post-approval activities have been well researched [11]. Previous studies have for instance identified mentions of ADRs from Twitter posts [36], used patient-health forums to detect drug–drug interactions [37] or identified patients switching from one treatment to another using a combination of online social data [38]. As for the available volume of social media data, previously utilised volumes range from a few hundred examples [39] to tens of thousands [12] or several hundred thousand or more [40]. Bian et al. [11] processed a total of 2 billion tweets, although not collected specifically for their study.
1.3 Drawbacks of Social Media for Health Research
At the same time, the utility of social media for patient health research remains debated [10]. For instance, although social media can offer unique insights into the patient perspective and patients are able to display their medical conditions with varying degrees of sophistication [12], the possibility to use social media for detecting new or early safety signals has been questioned [41]. On the one hand, prior studies have concluded that social media cannot identify new safety signals nor detect them earlier relative to other sources [42, 43]. On the other hand, there have been suggestions that uncommon or unknown AEs caused by drug–drug interactions could be discovered in Twitter posts [40]. In addition, in specific cases, certain safety signals may be detectable in advance. A 2022 study concluded that early signals of undesirable effects from Levothyrox use in France could be detected by mining patient comments from an online health forum [44]Footnote 1.
Furthermore, online users may not be representative of the full patient population [22], which is complicated by the difficulty in deriving online user demographics [45]. Active users also tend to be younger, are more likely to be women and are less acutely ill [46], whereas functionally impaired and less educated people are less likely to engage in Internet use [47]. Still, social media usage is increasing among older populations [48], and younger carers or relatives may engage in online discussions on behalf of older patients [49]. In addition, while undoubtedly problematic from a data privacy perspective, several studies have successfully derived user demographics using various online features such as search query history, user language or reposting behaviour [50,51,52,53], although this approach has been underutilised in the scope of mining social media for health information [45].
Social media have also been criticised for a lack of verifiable cause–effect relationships between drugs and AEs mentioned in posted content [54]. Compared with conventional drug safety reports, following up with patients to ask questions about their treatments and establish root causes behind events may be difficult if not impossible [55]. At the same time, technological advancements in NLP tools, with a demonstrated capability to distinguish between for example drug indications and AEs mentioned in social media [56] or the use of modern model architectures to detect posts with mentioned cause–effect relationships [57], unlock the possibility to detect suspected cause–effect relationships in social media, although definite conclusions on causality cannot be ascertained.
Last, regulatory acceptance of social media may not be on par with that of traditional sources [41], complicating its use. Health authorities enforce strict regulations as to how marketing authorisation holders, or companies authorised to market pharmaceutical products, should handle safety events. However, such regulations do not necessarily recognise the specifics of social media. Per FDA’s regulations, each safety event concerning a pharmaceutical company’s marketed drugs found in social media would have to be reported, resulting in the need for vast human resources [22].
Understanding this in light of the current evidence base for drug safety profiles as well as extant safety reporting methods is however necessary. For instance, scrutiny of conventional randomised clinical trials have revealed several of their shortcomings, such as isolated trial designs risking an under-representation of minorities [1]. Meanwhile, estimations suggest that up to 95% of treatment-related AEs outside of clinical trials remain undocumented by healthcare professionals [8, 58]. Furthermore, by definition, clinical trials do not capture AEs for off-label uses of drugs.
Systems such as the FDA’s Adverse Event Reporting System (FAERS) offer databases on suspected AEs and medication errors submitted to the health authority. FAERS has been a popular tool for deep learning-based studies in pharmacovigilance [59]. However, it has been noted to contain unspecified causal links, incomplete data or duplicated reporting, and under-reporting or over-reporting of known ADRs [35]. Thus, while the database contains reports on AEs and a particular product, this does not mean that the product caused the event [60]. In addition, evidence suggests sometimes considerable delays between safety events surfacing to pharmaceutical companies and the FDA receiving the information [61].
1.4 Regulatory Framework
1.4.1 RWD and Real-World Evidence (RWE)
The FDA defines real-world evidence (RWE) as “the clinical evidence about the usage and potential benefits or risks of a medical product derived from analysis of RWD”, and real-world data as “data relating to patient health status and/or the delivery of health care routinely collected from a variety of sources”, including electronic healthcare records, retrospective database studies, healthcare claims, social media, survey data and spontaneously reported AE data [3, 62]. In other words, RWD refers to data on patient health and RWE is the validated evidence derived from that data. Real world data can therefore be used to develop RWE; however, real-world data are not by default considered RWE.
Usage of RWD and RWE in a pre-approval setting remains relatively unexplored despite offering numerous opportunities for drug development. These include leveraging prescription data for identifying off-label drug use [63], using RWE for synthetic control arms (substituting a clinical trial’s control group using existing patient data sources) [9] or generating insights into the needs of a therapeutic area [62] , to name a few. Instead, the majority of applications so far have revolved around post-approval activities or support of orphan or life-threatening diseases [64, 65].
While RWD has been under-utilised in drug development, acknowledgment from regulatory bodies has also been limited. This is notable because regulatory guidance on how RWD can be used to support product approvals has been suggested as one of the most important factors for realising its full potential [2, 64]. A scrutiny of the regulatory frameworks as a proxy for understanding future use cases of RWD/RWE is therefore needed.
1.4.2 Regulatory Recognition
Pursuant to the 21st Century Cures Act, in 2018, the FDA released the Real-World Evidence Program to promote the use of RWE in regulatory decision-making processes [6]. The FDA’s Real-World Evidence Framework outlines how the FDA is expected to evaluate the use of RWE to support the approval of label expansions (i.e. broadening the number of medical conditions a drug can be prescribed for based on evidence from real-world usage), or phase IV study requirements (a post-approval clinical trial intended to study the drug’s effectiveness once already in public health use) [3]. The framework further covers definitions of RWD and RWE, use cases of RWD and examples of clinical trials using RWD/RWE, and plans for data standards [3, 6]. As regulatory maturity increases, a similar increase in usage of premarket applications of RWD/RWD may likewise be expected, propelled by their increasingly recognised utility.
For social media in particular, the FDA has released a series of guidance documents addressing how stakeholders can collect and submit patient experience data for drug development and regulatory decision making. In the first of its guidance documents, the agency highlighted social media, acknowledging its potential application in a development or regulatory setting: “targeted social media searches may be useful during the preliminary stages of a study to complement literature review findings, inform the development of research tools […], or as a supplement to traditional research approaches” [9]. The agency further outlines the strengths and limitations of using social media for gathering patient input, some of which have been noted in prior literature. Strengths include easy access to social media data, low burden for patients to provide the data and the potential for gathering information on health conditions. Limitations include the fact that participants are unknown, patient populations are not verifiably representative and the underlying selection process is difficult if not impossible to quantify. Furthermore, for an industry practitioner submitting information for a regulatory review, there is a need to include how the chosen data collection methods mitigate these limitations [9].
While not addressed specifically to social media data, a recent wave of regulatory guidance documents does highlight a rapid development of practical guidelines that industry practitioners can incorporate real-world findings in drug development and regulatory decision making [66]. This includes guidance documents [67] as well as the Agency’s Advancing Real-World Evidence Program [68], seeking to improve the quality and acceptability of RWE-based approaches to support labelling claims, including post-approval label expansions or to satisfy post-approval study requirements.
2 Methodology
We collected RWD from X by identifying drug names of interest, and then retrieving posts containing these drug names. A subset of the collected posts was annotated based on whether they contained a drug–effect relationship, and the annotated posts were used to train a machine learning model to automatically identify posts that contain a drug–effect relationship. The model was then used on the entire corpus of collected posts, and the presence of AEs was identified in those posts that contain that relationship. The quantitative data collection, processing and analysis was further informed by qualitative data collection, by means of interviews with industry professionals combined with participant observation conducted by one of the authors. The sections below provide a more detailed description of the methodology.
2.1 Search Query
We began by defining a list of generic anticoagulants (the chemical name of the product) and corresponding brand names (the given name by the producing company) to collect posts relating to the targeted drug class, anticoagulation. Anticoagulants were targeted based on the expectation that the volume of online discussions would be vast, given the size of annual spending dedicated to this therapeutic area [69]. Anticoagulants were also suggested during interviews with the collaborating company because they have been frequently discussed for their complexity in terms of for example dosages, foods interfering with the treatment and interactions with other drugs. Relevant generic names were identified through reviewing scientific literature on anticoagulation drugs [70, 71] and online providers of drug information such as Drugs.com. This resulted in a list of 14 generic names. The list of generic names was corroborated by a regulatory professional from the collaborating pharmaceutical company and five professionals from this company who work in the areas of regulatory affairs, pharmacovigilance and text mining.
Further, we identified 27 brand names from English-speaking countries and English-language posts (one product can have several names depending on the country or region). To identify brand names, we used the Physician’s Desk Reference, Drugs.com, the Merck Index and searched mentions of brand names across all MEDLINE articles from 2010 to 2020. The final list contained 34 keywords, including 14 generic and 20 brand names, outlined in Table 1. The “ATC class” column shows the subclass of anticoagulants per WHO’s Anatomical Therapeutic Chemical (ATC) codes. Accounting for possible misspellings of drugs was beyond the scope of this study, although we can add a heuristics-based approach [72] or entity normalisation [73] to address this issue in the future if our pipeline is moved into a production system.
2.2 Data Preparation
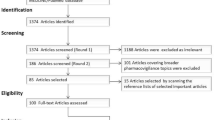
Data were collected using the social media analytics tool Sprinklr, offered by the collaborating company, which allows unlimited retrospective collection of X posts [74]. Posts were collected from 1 July to 31 December, 2019. Posts mentioning any of the 34 keywords were captured, while reposts and non-English posts were excluded. There were 14,993 posts collected (see Table 2).
We removed posts from users with over 5000 followers to exclude news outlets and celebrities [40, 54]. Usernames were replaced with numerical IDs and mentions of usernames in posts were removed using regular expressions to anonymise the dataset. The final dataset contained 10,264 posts.
2.2.1 Annotation
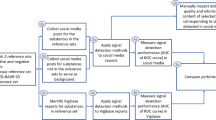
The annotation strategy is shown in Fig. 1. We followed a process similar to [13] and adapted it to our goal of labelling drug–effect relationships. Posts without a valid product mention were removed. Posts containing mentions of AEs but not a cause–effect relationship between a drug and an AE were labelled as negative, while posts containing both an AE and an indication that this was caused by a drug were labelled as positive. The aim of this multi-step approach to labelling was to specifically identify posts with a cause–effect relationship between a drug and AE, and not merely a co-occurrence of a drug name and AE.
Each post was annotated independently by each of the authors. Disagreements were resolved in a consensus meeting. Cohen’s Kappa for inter-rater agreement was 0.67.
In Table 3, the first three posts contain a specified effect suspected to be caused by a taken drug, while the latter three (a) mention a drug’s intended effect (such as for preventing blood clots), hence not being an AE, (b) represent an opinion or statement related to the drug, but not related to an AE, or (c) mention a drug and an event without specifying a relationship between the two.
We manually annotated 666 posts for the presence or absence of drug-caused AEs, with 466 negative instances and 200 positive instances (i.e. containing a relationship between a drug and an AE). The 70/30 distribution was chosen to reflect the natural distribution of the dataset [35]. This was further divided into two datasets: a training set of 500 instances (150 positive), and a test set of 166 instances (50 positive).
2.3 Machine Learning Modelling, Tuning and Prediction
For benchmark models, we used SVM, XGB and RF, with TF-IDF text vectorization. These were selected because of their light computational demand, proven performance in classification tasks [75] and popularity among researchers for classifying social media data [36, 54, 76]. Hyperparameter tuning was performed for the benchmark models using grid search (see Table 4).
These benchmark models were also combined into a single ensemble model with hard voting. That ensemble was used as a baseline model, to be compared with more powerful models, following common practice in machine learning.
We also experimented with more contemporary, transformer-based models. We chose ALBERT [77], which has lower computational requirements compared with most other transformer-based models. ALBERT’s base configuration has 12 layers of the transformer encoder [78], similar to BERT’s architecture [31]. Unlike BERT, however, ALBERT uses weight sharing between layers, leading to a significant reduction in the number of parameters in the model.
A pretrained ALBERT model was fine-tuned on the same training set of 500 instances as our benchmark models. For all models, performance was averaged across three runs with different train/test splits.
We focused on optimising model precision (also called PPV), which emphasises correct identification of drug-caused events among events marked by the model as containing an event. By contrast, optimising for recall (also called sensitivity) would emphasise capturing as many posts mentioning an AE as possible, potentially also capturing posts without AEs. A typical machine learning project will face a trade-off between increasing precision (thus decreasing the number of false positives) and increasing recall (decreasing the number of false negatives) [75]. As the focus in this study is on confidence in the accuracy of information found in posts, rather than finding more potentially relevant posts, we optimised for the precision metric. There were 1313 posts predicted as positive by the baseline model and were carried on in the data pipeline for further processing.
2.4 Text Mining: Medical Entity Extraction
I2e is a text-mining platform that uses a semi-supervised approach whereby users can interactively define pattern extraction rules for indexed text data [34]. We used I2e to identify and extract terms that describe AEs, diseases or disorders, foods or pharmacological substances, and translate these into medical terminology. Table 5 shows an example of the I2e output.
I2e returned 2020 assertions from the 1313 posts indexed into it. After deduplication, 1683 assertions across 685 unique posts were returned.
2.5 Industry Interviews and Participant Observations
The execution of the data processing pipeline and analysis was augmented by collecting qualitative input from industry professionals at a multinational pharmaceutical company via unstructured interviews. Outlined in Table 6, the unstructured interviews were carried out over 11 separate sessions with four professionals working across regulatory affairs, safety surveillance and information science.
In addition, one of the authors undertook a 6-month professional engagement with the pharmaceutical company during which continuous participant observations took place. Combined with the unstructured interviews, these brought further depth to the authors’ understanding of the pharmaceutical industry in general and key drug development processes in specific, and furthermore providing insights on developing the text mining pipeline using I2e as well as key factors influencing drug makers from a safety surveillance point of view when collecting social media data. The outcome of the interviews and observations is presented in the results.
3 Results
3.1 Machine Learning and Text Mining Results
The ALBERT model demonstrated higher recall (0.78) than the baseline model (0.66) and slightly higher precision (0.81 vs 0.79), with lower variability between models trained on different splits of training and test data, as demonstrated by the lower standard deviation (see Table 6).
The 1313 predicted posts indexed into I2e and mined for drug events produced 1683 total hits and 1182 unique hits across 685 posts, posted by 597 unique users (see Table 7). It follows that some posts comprised multiple findings and that while some hits were duplicated, I2e identified several entities within the same hits (Table 8).
The most frequently mentioned drugs were warfarin and drugs within the direct factor Xa inhibitors class: betrixaban, edoxaban, apixaban and rivaroxaban (Fig. 2). Three drugs had zero matches in the I2e query (argatroban, desirudin, danaparoid). Among drugs mentioned in addition to the anticoagulants, different types of pain relievers/anti-inflammatory drugs had the greatest occurrence (Fig. 3). Various forms of haemorrhage, fatal drug outcomes, stroke, and different types of pains or bruises were the most frequently mentioned drug events across all posts, with the former three comprising more serious ADRs (Fig. 4). Stroke, notably, can both be prevented by anticoagulation therapy (in the case of ischaemic stroke) or be a complication of it (in the case of intracranial haemorrhage) [79].
Per Fig. 3, the combined terminologies in I2e proved to produce some overlap as the pain relievers aspirin and ketorolac are included in the drug class NSAIDs (non-steroidal anti-inflammatory drugs) [80]. Therefore, aspirin, ketorolac and NSAIDs were grouped together under the label “NSAIDs” for subsequent visualisations. The most common ADRs and AEs in Fig. 3 concur with current knowledge and include haemorrhage, death or stroke. Adverse effects involving various forms of pains or bruises are established events within anticoagulation and/or common reports in FAERS [70, 71]. Notably, the number of unique users mentioning each event also remained high in all cases, despite being lower than the total count of events. This suggests that these events are widely dispersed among users, even though some users mention an event several times.
Warfarin was over-represented in mentions of the most common drug events (Fig. 5). This distribution remained similar when comparing the frequency across drug classes instead of drugs except for mentions of haemorrhage, which was more equally represented within the classes vitamin K antagonists (i.e. warfarin) and direct factor Xa inhibitors.
3.2 Industry Observations
Interviews with industry professionals provided several insights on the role of RWD and social media in conventional drug development. First, clinical development should not be considered solely as the precedent for developing the drug label (i.e. the drug’s prescribing information). Rather, the two components (clinical development and producing the prescribing information) should iteratively inform each other. For instance, research during labelling activities also provides guidance on clinical trial design to optimise the safety and efficacy profile of the drug, based on input from multiple sources of regulatory intelligence such as competitors’ labels or the clinical trials that support them. Capturing the right data and intelligence, outside of traditional randomised clinical trials, is therefore a key component of regulatory decision making and drug development.
Second, for social media in particular, currently there is little industry precedent for its use: social media has so far not been widely used in regulatory decision making, for deriving RWD, or for supporting regulatory or safety functions. For future usage, two potential use cases can be highlighted: (1) when a drug developer has no drug on the market within the area of interest, competitors’ drugs could be targeted for understanding what online health discussions were considering important for drugs in the given area and (2) conversely, when there is a drug marketed, social media can be used to collect and analyse health discussions for understanding either how that drug compared with other similar drugs, or for getting a greater general understanding of the patient perspective.
Third, multiple challenges around social media-derived RWD were emphasised. Regulatory recognition of RWD, albeit maturing, was still seen to present considerable complexities in terms of collection, analysis and integration of real-world findings with clinical development, including in defining the role of RWD as a supplement to the existing evidence base. In addition, the availability of RWD remained a challenge, with currently undefined processes for how to obtain and manage sources of RWD.
Finally, the lack of opportunities to infer causality or validate findings from social media was considered less of a challenge than currently conceptualised in the literature. Instead, social media, and experimental sources of RWD in general, were perceived positively in terms of their possibility to generate hypotheses on areas to be investigated further.
4 Analysis and Discussion
4.1 Positioning Social Media Data in the Drug Development Toolbox
Extant research and our results position social media as offering direct access to the patient experience. However, establishing causality or validating any findings suggesting causality remains difficult. Meanwhile, industry practitioners suggest that it is not only evidence of treatment outcomes that is valuable in the context of drug development. Capturing external intelligence, outside of clinical development, that better informs the development process is also considered important. For social media to serve this purpose, it needs to generate novel and actionable insights, and in addition meet these criteria to the extent where they can actually support the drug development process. To evaluate social media against these criteria, we explore specific use cases.
4.2 Use Cases for Social Media-Derived RWD
Our data suggest that social media may paint a more holistic picture of patients’ health conditions, i.e. what a typical patient experiences in a given therapeutic area looks like. Building on this, Fig. 6 shows the most frequent findings mentioned in addition to haemorrhage or stroke respectively. It shows, for all patients mentioning haemorrhage (left side) or stroke (right side) in a post, the most common other findings mentioned by those same users.
Figure 6 shows that haemorrhage appears to be one of the main concerns for patients discussing experiences relating to stroke. In discussions of haemorrhage, death or fatal outcomes, atrial fibrillation (irregular heartbeat) and stroke all appear as the most frequent mentions. In addition, for the haemorrhage group, blood clots, pains or neoplasms (tumours) seem to be adverse effects present for many patients, albeit less frequent. Meanwhile, renal failure (various kidney issues) appears to be more common among discussions on haemorrhage than those on stroke.
While these results provide some insight into the patient experience, the findings still need to be both new and actionable in order to support the drug development process. We argue that social media may do this by supporting the generation of hypotheses.
First, in the data, we observe that patients who mention stroke or haemorrhage as AEs appear to have concomitant diseases (multiple co-existing diseases). Specifically, renal failures and atrial fibrillations are mentioned in both groups, and more frequently in the haemorrhage group. Second, many patients mentioning haemorrhage also mention neoplasms; however, neoplasms are not mentioned across patients mentioning stroke. Potential hypotheses could be as follows: (1) renal failure or atrial fibrillation are more prevalent conditions in patients receiving anticoagulation therapy experiencing haemorrhage events, than in those experiencing a stroke and (2) there is a connection between patients experiencing haemorrhage events and an increase in the risk of neoplasm or potential tumours, or vice versa.
Second, if these hypotheses are rejected through follow-up tests or past data on adverse effects, this may in itself be valuable information. Discussions of non-existing adverse effects highlight potential knowledge gaps in the patient population, making it possible to formulate hypotheses about beliefs. The corresponding hypotheses to the two outlined above, focusing on beliefs instead of outcomes, could be as follows: (1) patients prescribed anticoagulants are erroneously concerned with a greater risk of neoplasm as an effect of taking blood thinners and (2) current prescription information available to patients prescribed anticoagulants for atrial fibrillation does not communicate the associated risks of haemorrhage well enough. This is actionable information that drug makers or regulators can use to improve labelling or otherwise better inform the patient population.
Last, although the question remains whether these are new findings—as these hypotheses may only relate to known information—such findings can still confirm an already expected reality. If no new events occur in online health discussions, that could serve to further indicate that all potential events are already accounted for. That is, even a non-finding should be considered a finding, and given the low use of social media in gauging patient experiences to date, the medium arguable offers novel insights.
Still, presuming that social media can (a) provide novel insights into the patient experience and (b) that these insights can be operationalised by means of generating hypotheses, it also needs to be proven both feasible and valuable enough to support the drug development process. We suggest three possible scenarios to support this process.
Scenario A: suspected new safety events are detected in social media. Drug makers can use these indications to guide the generation of measurable hypotheses and include them as endpoints in clinical trials, or guide one-on-one patient interviews.
Scenario B: suspected misbeliefs about possible drug–event relationships are noticed. Drug makers can address such misbeliefs in the medication guidance to patients, or inform regulators that knowledge gaps exist among patients within a given therapeutic area. A dialogue on how to address these gaps can ensue, for example, via supporting healthcare providers with better education materials.
Scenario C: no new events are detected in social media. Drug makers would conclude with a higher likelihood that extant knowledge of the drug covers all possible AEs and safety events. This finding could be included in submissions to regulators as an additional measure that has been taken to ensure proper prescribing information. This also includes situations when events discussed on social media form a subset of events observed in prior trials. In addition, the prevalence of mentions of already known (mis)beliefs in social media can also indicate which factors are considered more, or less, critical to patients. For example, in the case of off-label use where (albeit already known or suspected that patients may be erroneously using a drug outside of its approved indication) social media can serve as an additional source to better understand the magnitude of the issues at hand, guiding experts towards the most important topics.
To conclude Sect. 4.2, we argue that social media can efficiently enhance the drug development process. The medium can offer novel insights even when those insights only serve to corroborate an already suspected reality, allowing further guidance to experts who would be able to translate these insights into action. For example, coronavirus disease 2019 vaccines, which were developed and assessed within a year and then distributed to vast populations globally, were surrounded by misinformation and public misconceptions about efficacy and adverse effects [81]. In such a scenario, social media may be more likely to capture events, perspectives or relationships from real-world experiences that are not yet identified in extant clinical trials. While not targeted in this specific study, analysis of social media data may also reveal common conditions for which specific drugs are used off-label [28, 29], as well as shed light on patient outcomes and awareness of risks and benefits of off-label use. This will enrich the assessment of risks with off-label drug use by the regulators with the patient perspective.
4.3 Social Media Usage and the Regulatory Landscape
Considering the FDA’s 2020 guidance on social media usage in patient-focused drug development [9], there is increasing regulatory recognition of the above proposed use cases. Social media provides an opportunity to surface potential concomitant diseases, patient beliefs about potential drug–event relationships or other health issues experienced by patients. These insights could supplement drug developers’ or regulators’ understanding of the patient population and typical health conditions or guide one-on-one interviews with patients conducted as part of a clinical trial. These findings could be argued to align with FDA’s envisioned usage of social media (see Sect. 1.4.2), making it plausible to situate social media-derived RWD in the current regulatory landscape.
Such usages are likely to come with regulatory requirements that need to be met. For instance, the FDA guidance also notes that when submitting information for a regulatory review, the sponsor should demonstrate how data collection addressed the limitations in social media, such as the lack of opportunities to verify patient identities [9]. It may not be clear what falls under the scope of this requirement. If social media are only used to generate hypotheses that are then investigated and verified in traditional clinical development, do social media constitute part of the evidence base and thus need to be included for a regulatory review? In addition, one of the findings of this study is that multiple drug events or disorders posted by a single user can be connected to that user, even when their data are anonymised. While beneficial for understanding drug interactions and comprehensive health conditions, gathering such information raises potential privacy issues [24], as even anonymised data can be susceptible to privacy attacks [82]. With little precedent, it is currently difficult to conclude on the precise implications of these requirements.
4.4 Implications for Industry
This paper incorporates several key themes for leveraging social media for RWD in a regulatory, drug development setting: social media for health information, the regulatory landscape for RWD and machine learning and NLP. This allows us to provide developers with a more exhaustive overview of how social media can be used in practice, while also offering new perspectives on some of its previously mentioned shortcomings. The use case suggests how social media can supplement the traditional drug developer’s toolbox, as a potential tool for generating hypotheses to be further investigated via extant methods in clinical development. By situating social media in the drug development stage rather than only post-approval, the often-scrutinised lack of validated evidence in social media data appears less important to support clinical development. In addition, while current works aim to develop more advanced approaches for distinguishing between the types of medical entities encountered in social media, such as ADRs and indications [56], by targeting a single drug class, this study instead highlighted the possibility to draw on existing knowledge of that class and therefore make the distinction between the different types of entities.
Industry actors still need to be wary of social media usage. On the one hand, imposed safety requirements complicate usage from an operational point of view. On the other hand, from a regulatory point of view, the regulations are not yet clearly defined and little precedence for incorporating social media in clinical development exists. Notably, the FDA released their second guidance document on patient-focused drug development in February 2022, more than 1.5 years after the first guidance, titled “Methods to Identify What Is Important to Patients” [83]. While some new general guidance on social media usage is included, defined use cases and regulatory frameworks for social media data mining pipelines remain unspecified. Furthermore, because social media users may have a different demographic than the general population, social media monitoring should be complemented with other approaches from the practitioner’s toolbox.
The use cases suggested in the industry observations are considered to remain valid in light of our results and analysis. In the drug development phase, a pharmaceutical company can use social media for gathering RWD on competitors’ already launched treatments to better understand the patient experience and, thereby, consider whether to account for such insights in clinical trials. Or, in the event where a company has a drug launched, social media can be used to inform subsequent post-approval clinical trials, or the development of additional educational material to patients or healthcare providers. In addition, social media can be used to provide insight into the outcomes of drugs that are used off-label [28, 29].
4.5 Implications for Regulators
This paper sheds further light on the width of the gap between technology and regulations. While data from social media and the technology for its analysis appear mature enough to warrant, at a minimum, exploratory usage in industry, regulatory maturity appears to be lagging behind. In practice, further guidance is needed on the role of social media in the pre-approval stage of drug development. Such guidance should account for the operational challenges currently faced by drug developers in incorporating social media as a source of RWD. Considering the current lack of precedent, regulators should also translate potential use cases into actionable regulatory frameworks or consider how novel concepts, such as regulatory sandboxes, can encourage exploratory use.
4.6 NLP and Health Information Mining
The results of NLP modelling demonstrate an improvement compared with prior work [40, 54]. In particular, ALBERT outperformed the ensemble baseline model, while requiring substantially less hyperparameter tuning and data preprocessing. This made the ALBERT model significantly easier to use compared with the baseline model, which did include all of these steps. Because we fine-tuned a model that had been previously pretrained, we were able to achieve these better results while using a modest-sized dataset. This means that practitioners are recommended to use this transfer learning approach, when language models previously pre-trained on large datasets are fine-tuned on smaller datasets for the task at hand (e.g. text classification), as it may yield a better result with a lower amount of time spent on modelling. The potential downside of this approach, however, is the need for performant hardware, including a graphical processing unit. At the same time, cloud-based hardware can be used, with appropriate privacy measures in place where necessary when dealing with sensitive or personally identifiable information.
The posts finally outputted reveal potential to access patient health insights on, for example, adverse drug events or therapeutic area pain points by mining social media. Most notably, relative user diversity as measured by the percentage of posts from unique users out of all posts increased as data moved through the data pipeline. The 14,993 posts initially collected were posted by 8573 unique users. Through data preprocessing, supervised classification and the text mining query, the number of unique users relative to the total number of posts increased, as seen in Table 9. Thus, while the number of posts decreased significantly, user diversity increased, thus offering a broader understanding of the patient perspective.
Further, our pipeline resulted in a model that should capture cause–effect relationships between drugs and AEs. This is because our annotation scheme aimed to capture only posts describing a relationship between a drug and an AE, and posts with simple co-occurrences of drug names and adverse effects (but no cause–effect relationship) were assigned a negative label. This allowed us to incorporate recent work on detecting cause–effect relationships in posts [57] as part of the overall pipeline of using social media for RWD.
5 Conclusions
This paper develops a data pipeline for extracting health information from Twitter posts on anticoagulants, investigates the applicable regulatory landscape for social media and RWD, and situates these findings in industry practice. We conclude that RWD derived from social media shows potential for supporting regulatory decision making for pharmaceutical companies, by generating hypotheses on patient conditions, experiences and beliefs. Ultimately, this can aid conventional clinical development.
Some recognition is found amongst regulators. However, the immaturity of social-media specific regulations and, therefore, the lack of precedent for how findings would be perceived under a regulatory review, mean that the role of social media as a supplementary source of information remains to be precisely defined.
While technological challenges persist, specifically those relating to deriving cause–effect relationships, we suggest this is less of an issue than previously argued. To the best of our knowledge, this paper is the first to conclude that how you use the medium may overcome some of its argued shortcomings, i.e. utilising social media to generate hypotheses, not evidence.
This study is limited by its use of posts written only in English, which narrows the scope of information captured. Similarly, using only X data means omitting potential insights shared by patients in other online forums. Furthermore, language cannot determine region specificity, complicating scenarios where approved indications and off-label prescription habits of healthcare providers may differ between countries. Last, the paper uses the baseline model to make predictions which in turn inform subsequent data analysis steps. While this does not allow us to benchmark results, the purpose of this paper is exploratory and to develop a pipeline, which it accomplishes.
Future studies may expand on this paper by combining several online sources written in different languages, and by changing components of this data pipeline and benchmarking the results against ours. Use of social media in pre-approval drug development, such as informing clinical trials, is a particularly under-researched area, where the hypotheses generation approach described in this paper may yield fruitful results. This study can also inform future research on reconciling technological developments with health and safety regulations.
Change history
14 June 2024
A Correction to this paper has been published: https://doi.org/10.1007/s40264-024-01459-9
Notes
We thank an anonymous reviewer for suggesting this case.
References
Hoppe C, Kerr D. Minority underrepresentation in cardiovascular outcome trials for type 2 diabetes. Lancet Diabetes Endocrinol. 2017;5:13.
Morgan J, Feghali K, Shah S, Miranda W. RWE focus is shifting to R&D, early investments begin to pay off. How can others catch up? 2020. https://www2.deloitte.com/content/dam/insights/us/articles/6578_CHS-RWE-benchmarking-survey/DI_RWE%20benchmarking%20survey%20(SECURED).pdf. Accessed 21 Feb 2024.
US FDA. Framework for FDA’s real world evidence program. 2018. Available from: https://www.fda.gov/media/120060/download. Accessed 24 Jan 2023.
Shen N-N, Zhang C, Hang Y, Li Z, Kong L-C, Wang N, et al. Real-world prevalence of direct oral anticoagulant off-label doses in atrial fibrillation: an epidemiological meta-analysis. Front Pharmacol. 2021;12: 581293.
The Economist. How health care is turning into a consumer product. 2022. https://www.economist.com/business/how-health-care-is-turning-into-a-consumer-product/21807114. Accessed 29 Dec 2022.
Klonoff DC, Gutierrez A, Fleming A, Kerr D. Real-world evidence should be used in regulatory decisions about new pharmaceutical and medical device products for diabetes. J Diabetes Sci Technol. Los Angeles (CA) SAGE Publications; 2019: p. 995–1000.
Hiki N, Honda M, Etoh T, Yoshida K, Kodera Y, Kakeji Y, et al. Higher incidence of pancreatic fistula in laparoscopic gastrectomy: real-world evidence from a nationwide prospective cohort study. Gastr Cancer. 2018;21:162–70.
García-Abeijon P, Costa C, Taracido M, Herdeiro MT, Torre C, Figueiras A. Factors associated with underreporting of adverse drug reactions by health care professionals: a systematic review update. Drug Saf. 2023;46(7):625–36.
US FDA. Patient-focused drug development: collecting comprehensive and representative Input. guidance for industry, Food and Drug Administration staff, and other stakeholders. 2020. https://www.fda.gov/media/139088/download. Accessed 24 Jan 2023.
Pappa D, Stergioulas LK. Harnessing social media data for pharmacovigilance: a review of current state of the art, challenges and future directions. Int J Data Sci Anal. 2019;8:113–35.
Bian J, Topaloglu U, Yu F. Towards large-scale Twitter mining for drug-related adverse events. SHB12 (2012). 2012;2012:25–32.
Freifeld CC, Brownstein JS, Menone CM, Bao W, Filice R, Kass-Hout T, et al. Digital drug safety surveillance: monitoring pharmaceutical products in Twitter. Drug Saf. 2014;37:343–50.
Powell GE, Seifert HA, Reblin T, Burstein PJ, Blowers J, Menius JA, et al. Social media listening for routine post-marketing safety surveillance. Drug Saf. 2016;39:443–54.
Lardon J, Abdellaoui R, Bellet F, Asfari H, Souvignet J, Texier N, et al. Adverse drug reaction identification and extraction in social media: a scoping review. J Med Internet Res. 2015;17: e4304.
Vilar S, Friedman C, Hripcsak G. Detection of drug–drug interactions through data mining studies using clinical sources, scientific literature and social media. Brief Bioinform. 2018;19:863–77.
Buyuk SK, Imamoglu T. Instagram as a social media tool about orthognathic surgery. Health Promot Perspect. 2019;9:319–22.
Dai H-J, Wang C-K. Classifying adverse drug reactions from imbalanced twitter data. Int J Med Inf. 2019;129:122–32.
Jordan S, Hovet S, Fung I. Using Twitter for public health surveillance from monitoring and prediction to public response. Data. 2019;4:6.
White E, Read J, Julo S. The role of Facebook groups in the management and raising of awareness of antidepressant withdrawal: is social media filling the void left by health services? Ther Adv Psychopharmacol. 2021;11:204512532098117.
Dreisbach C, Koleck TA, Bourne PE, Bakken S. A systematic review of natural language processing and text mining of symptoms from electronic patient-authored text data. Int J Med Inf. 2019;125:37–46.
Fox S, Duggan M. Health online 2013. 2013. https://www.pewresearch.org/internet/2013/01/15/health-online-2013/. Accessed 29 Dec 2022.
Sloane R, Osanlou O, Lewis D, Bollegala D, Maskell S, Pirmohamed M. Social media and pharmacovigilance: a review of the opportunities and challenges: social media and pharmacovigilance. Br J Clin Pharmacol. 2015;80:910–20.
Okon E, Rachakonda V, Hong HJ, Callison-Burch C, Lipoff JB. Natural language processing of Reddit data to evaluate dermatology patient experiences and therapeutics. J Am Acad Dermatol. 2020;83:803–8.
Gupta A, Katarya R. Social media based surveillance systems for healthcare using machine learning: a systematic review. J Biomed Inform. 2020;108: 103500.
Mithani Z. Informed consent for off-label use of prescription medications. AMA J Ethics. 2012;14:576–81.
Han SH, Safeek R, Ockerman K, Trieu N, Mars P, Klenke A, et al. Public interest in the off-label use of glucagon-like peptide 1 agonists (Ozempic) for cosmetic weight loss: a Google Trends analysis. Aesthet Surg J. 2024;44:60–7.
Hua Y, Jiang H, Lin S, Yang J, Plasek JM, Bates DW, et al. Using Twitter data to understand public perceptions of approved versus off-label use for COVID-19-related medications. J Am Med Inform Assoc. 2022;29:1668–78.
Duguay S. You can’t use this app for that: Exploring off-label use through an investigation of Tinder. Inf Soc. 2020;36:30–42.
Avram S, Halip L, Curpan R, Borota A, Bora A, Oprea TI. Annotating off-label drug usage from unconventional sources. MedRxiv Prepr medRxiv:2022.09.08.22279709. 2022.
Segura-Bedmar I, Martínez Fernández P. Pharmacovigilance through the development of text mining and natural language processing techniques. J Biomed Inform. 2015;58:288–91.
Devlin J, Chang M-W, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. ArXiv Prepr ArXiv181004805. 2018.
Casola S, Lavelli A. FBK@ SMM4H2020: RoBERTa for detecting medications on Twitter. Proc Fifth Soc Media Min Health Appl Workshop Shar Task; 2020: p. 101–3.
Shen Y, Heacock L, Elias J, Hentel KD, Reig B, Shih G, et al. ChatGPT and other large language models are double-edged swords. Radiology. 2023;307: e230163.
Ly T, Pamer C, Dang O, Brajovic S, Haider S, Botsis T, et al. Evaluation of Natural Language Processing (NLP) systems to annotate drug product labeling with MedDRA terminology. J Biomed Inform. 2018;83:73–86.
Sarker A, Ginn R, Nikfarjam A, O’Connor K, Smith K, Jayaraman S, et al. Utilizing social media data for pharmacovigilance: a review. J Biomed Inform. 2015;54:202–12.
Ginn R, Pimpalkhute P, Nikfarjam A, Patki A, O’Connor K, Sarker A, et al. Mining Twitter for adverse drug reaction mentions: a corpus and classification benchmark. Proc Fourth Workshop Build Eval Resour Health Biomed Text Process; 2014: p. 1–8.
Yang H, Yang CC. Harnessing social media for drug-drug interactions detection. 2013 IEEE Int Conf Healthc Inform. Philadelphia (PA): IEEE; 2013: p. 22–9. http://ieeexplore.ieee.org/document/6680457/. Accessed 29 Dec 2022.
Risson V, Saini D, Bonzani I, Huisman A, Olson M. Patterns of treatment switching in multiple sclerosis therapies in US patients active on social media: application of social media content analysis to health outcomes research. J Med Internet Res. 2016;18: e62.
Jiang K, Zheng Y. Mining Twitter data for potential drug effects. In: Motoda H, Wu Z, Cao L, Zaiane O, Yao M, Wang W, editors. Adv Data Min Appl. Berlin, Heidelberg: Springer Berlin, Heidelberg; 2013: p. 434–43. https://doi.org/10.1007/978-3-642-53914-5_37. Accessed 29 Dec 2022.
Hsu D, Moh M, Moh T-S. Mining frequency of drug side effects over a large twitter dataset using apache spark. 2017 IEEEACM Int Conf Adv Soc Netw Anal Min ASONAM. IEEE; 2017: p. 915–24
European Network of Centres for Pharmacoepidemiology and Pharmacovigilance. Guide on methodological standards in pharmacoepidemiology (Revision 8). 2020. Available from: https://www.encepp.eu/standards_and_guidances/documents/GuideMethodRev8.pdf. [Accessed 29 Dec 2022].
Bhattacharya M, Snyder S, Malin M, Truffa MM, Marinic S, Engelmann R, et al. Using social media data in routine pharmacovigilance: a pilot study to identify safety signals and patient perspectives. Pharm Med. 2017;31:167–74.
Caster O, Dietrich J, Kürzinger M-L, Lerch M, Maskell S, Norén GN, et al. Assessment of the utility of social media for broad-ranging statistical signal detection in pharmacovigilance: results from the WEB-RADR Project. Drug Saf. 2018;41:1355–69.
Roche V, Robert J-P, Salam H. AI-based approach for safety signals detection from social networks: application to the Levothyrox scandal in 2017 on Doctissimo Forum. ArXiv Prepr ArXiv220303538. 2022.
Sinnenberg L, Buttenheim AM, Padrez K, Mancheno C, Ungar L, Merchant RM. Twitter as a tool for health research: a systematic review. Am J Public Health. 2017;107:e1-8.
Padrez KA, Ungar L, Schwartz HA, Smith RJ, Hill S, Antanavicius T, et al. Linking social media and medical record data: a study of adults presenting to an academic, urban emergency department. BMJ Qual Saf. 2016;25:414–23.
Greysen SR, Chin Garcia C, Sudore RL, Cenzer IS, Covinsky KE. Functional impairment and internet use among older adults: implications for meaningful use of patient portals. JAMA Intern Med. 2014;174:1188.
Pew Research Center. Social media use in 2021. 2021. https://www.pewresearch.org/internet/2021/04/07/social-media-use-in-2021/. Accessed 29 Dec 2022.
McDonald L, Malcolm B, Ramagopalan S, Syrad H. Real-world data and the patient perspective: the PROmise of social media? BMC Med. 2019;17:11.
Bi B, Shokouhi M, Kosinski M, Graepel T. Inferring the demographics of search users: social data meets search queries. Proc 22nd Int Conf World Wide Web - WWW 13. Rio de Janeiro: ACM Press; 2013: p. 131–40. http://dl.acm.org/citation.cfm?doid=2488388.2488401. Accessed 29 Dec 2022.
Li D, Li Y, Ji W. Gender identification via reposting behaviors in social media. IEEE Access. 2018;6:2879–88.
Pennacchiotti M, Popescu A-M. Democrats, republicans and starbucks afficionados: user classification in twitter. Proc 17th ACM SIGKDD Int Conf Knowl Discov Data Min - KDD 11. San Diego (CA): ACM Press; 2011: p. 430. http://dl.acm.org/citation.cfm?doid=2020408.2020477. Accessed 29 Dec 2022.
Rao D, Yarowsky D, Shreevats A, Gupta M. Classifying latent user attributes in twitter. Proc 2nd Int Workshop Search Min User-Gener Contents - SMUC 10. Toronto (ON): ACM Press; 2010: p. 37. http://portal.acm.org/citation.cfm?doid=1871985.1871993. Accessed 29 Dec 2022.
Yu F, Moh M, Moh T-S. Towards extracting drug-effect relation from Twitter: a supervised learning approach. 2016 IEEE 2nd Int Conf Big Data Secur Cloud BigDataSecurity IEEE Int Conf High Perform Smart Comput HPSC IEEE Int Conf Intell Data Secur IDS. New York (NY): IEEE; 2016: p. 339–44. http://ieeexplore.ieee.org/document/7502313/. Accessed 29 Dec 2022.
Franzen W. Can social media benefit drug safety? Drug Saf. 2011;34:793.
Chowdhury S, Zhang C, Yu PS. Multi-Task Pharmacovigilance mining from social media posts. Proc 2018 World Wide Web Conf World Wide Web - WWW 18. Lyon, France: ACM Press; 2018: p. 117–26. http://dl.acm.org/citation.cfm?doid=3178876.3186053. Accessed 29 Dec 2022.
Ahne A, Khetan V, Tannier X, Rizvi MIH, Czernichow T, Orchard F, et al. Extraction of explicit and implicit cause-effect relationships in patient-reported diabetes-related tweets from 2017 to 2021: deep learning approach. JMIR Med Inform. 2022;10: e37201.
Hazell L, Shakir SA. Under-reporting of adverse drug reactions. Drug Saf. 2006;29:385–96.
Kompa B, Hakim JB, Palepu A, Kompa KG, Smith M, Bain PA, et al. Artificial intelligence based on machine learning in pharmacovigilance: a scoping review. Drug Saf. 2022;45:477–91.
FDA. FDA Adverse Event Reporting System (FAERS) public dashboard. 2021. https://www.fda.gov/drugs/questions-and-answers-fdas-adverse-event-reporting-system-faers/fda-adverse-event-reporting-system-faers-public-dashboard. Accessed 15 Oct 2023.
Ma P, Marinovic I, Karaca-Mandic P. Drug manufacturers’ delayed disclosure of serious and unexpected adverse events to the US Food and Drug Administration. JAMA Intern Med. 2015;175:1565.
Lamberti MJ, Kubick W, Awatin J, McCormick J, Carroll J, Getz K. The use of real-world evidence and data in clinical research and postapproval safety studies. Ther Innov Regul Sci. 2018;52:778–83.
Mezher M. Real world evidence: can it support new indications, label expansions? 2016. https://www.raps.org/regulatory-focus%E2%84%A2/news-articles/2016/3/real-world-evidence-can-it-support-new-indications,-label-expansions. Accessed 29 Dec 2022.
Bipartisan Policy Center. Using real-world evidence to accelerate safe and effective cures: advancing medical innovation for a healthier America. 2016. https://bipartisanpolicy.org/download/?file=/wp-content/uploads/2019/03/BPC-Health-Innovation-Safe-Effective-Cures.pdf. Accessed 29 Dec 2022.
Zou KH, Li JZ, Imperato J, Potkar CN, Sethi N, Edwards J, et al. Harnessing real-world data for regulatory use and applying innovative applications. J Multidiscip Healthc. 2020;13:671–9.
Burns L, Le Roux N, Kalesnik-Orszulak R, Christian J, Dudinak J, Rockhold F, et al. Real-world evidence for regulatory decision-making: updated guidance from around the world. Front Med. 2023;10:1236462.
US FDA. Considerations for the use of real-world data and real-world evidence to support regulatory decision-making for drug and biological products: guidance for industry. 2023. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/considerations-use-real-world-data-and-real-world-evidence-support-regulatory-decision-making-drug. Accessed 29 Dec 2023.
US FDA. Advancing real-world evidence program. 2023. https://www.fda.gov/drugs/development-resources/advancing-real-world-evidence-program. Accessed 29 Dec 2023.
Mikulic M. Leading 10 U.S. therapy areas based on drug spending in 2020. 2023. Available from: https://www.statista.com/statistics/238698/us-health-spending-leading-areas. Accessed 23 Oct 2023.
Harter K, Levine M, Henderson S. Anticoagulation drug therapy: a review. West J Emerg Med. 2015;16:11–7.
Myers K, Lyden A. A review on the new and old anticoagulants. Orthop Nurs. 2019;38:43–52.
Pimpalkhute P, Patki A, Nikfarjam A, Gonzalez G. Phonetic spelling filter for keyword selection in drug mention mining from social media. AMIA Summits Transl Sci Proc. 2014;2014:90.
Ji Z, Wei Q, Xu H. Bert-based ranking for biomedical entity normalization. AMIA Summits Transl Sci Proc. 2020;2020:269.
Sprinklr. Twitter as a Listening source. 2023. https://www.sprinklr.com/help/articles/twitter/twitter-as-a-listening-source/641c27fe55c4c33ae8b8149c. Accessed 15 Oct 2023.
Géron A. Hands-on machine learning with Scikit-Learn, Keras, and TensorFlow. Sebastopol, CA: O’Reilly Media, Inc.; 2022.
Wu L, Moh T-S, Khuri N. Twitter opinion mining for adverse drug reactions. 2015 IEEE Int Conf Big Data Big Data. Santa Clara (CA): IEEE; 2015: p. 1570–4. http://ieeexplore.ieee.org/document/7363922/. Accessed 29 Dec 2022.
Lan Z, Chen M, Goodman S, Gimpel K, Sharma P, Soricut R. ALBERT: A Lite BERT for self-supervised learning of language representations. arXiv; 2019. https://arxiv.org/abs/1909.11942. Accessed 21 Feb 2024.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Adv Neural Inf Process Syst. Long Beach, CA; 2017. p. 6000–10.
Staerk L, Fosbøl EL, Lip GYH, Lamberts M, Bonde AN, Torp-Pedersen C, et al. Ischaemic and haemorrhagic stroke associated with non-vitamin K antagonist oral anticoagulants and warfarin use in patients with atrial fibrillation: a nationwide cohort study. Eur Heart J. 2017;38:907–15.
WHO Collaborating Center for Drug Statistics Methodology. ATC/DDD Index. 2022. Available from: https://www.whocc.no/atc_ddd_index/. Accessed 29 Dec 2022]
Pierri F, DeVerna MR, Yang K-C, Axelrod D, Bryden J, Menczer F. One year of COVID-19 vaccine misinformation on Twitter: longitudinal study. J Med Internet Res. 2023;25: e42227.
Olatunji IE, Rauch J, Katzensteiner M, Khosla M. A review of anonymization for healthcare data. Big data. 2022. https://doi.org/10.1089/big.2021.0169. Accessed 21 Feb 2024.
US FDA. Patient-focused drug development: methods to identify what is important to patients. 2022. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/patient-focused-drug-development-methods-identify-what-important-patients. Accessed 29 Dec 2022.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
Open access funding provided by Copenhagen Business School.
Conflicts of interest/competing interests
Didrik Wessel is a current full-time employee at Novo Nordisk. Nicolai Pogrebnyakov was a full-time employee at Twitter after the research was completed but before the manuscript was published. Nicolai Pogrebnyakov is a current full-time employee at Sparrow Bioacoustics, a medical software company.
Ethics approval
Not applicable. This article represents a retrospective study conducted on publicly available data, which was anonymised immediately after data collection. No personal or personally identifiable data are included in the paper or any supporting materials.
Consent for publication
Not applicable.
Consent to participate
Not applicable.
Availability of data and material
The dataset supporting the conclusions of this article is not publicly available. The data consist of posts describing symptoms of health conditions. Even if these posts are anonymised by replacing usernames with unique identifiers, they could potentially be reverse engineered to identify individuals. To access the data, please e-mail the corresponding author.
Code availability
The code used in the analysis is available at https://github.com/nick-edu/rwd-tweets.
Authors’ contributions
DW collected the data, conducted interviews, performed participant observation, wrote the code and trained the baseline models. NP wrote the code and trained the ALBERT model and provided research guidance. Both authors labelled the data and wrote the paper. All authors read and approved the final version.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Wessel, D., Pogrebnyakov, N. Using Social Media as a Source of Real-World Data for Pharmaceutical Drug Development and Regulatory Decision Making. Drug Saf 47, 495–511 (2024). https://doi.org/10.1007/s40264-024-01409-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40264-024-01409-5