Abstract
Introduction
A large number of studies on systems to detect and sometimes normalize adverse events (AEs) in social media have been published, but evidence of their practical utility is scarce. This raises the question of the transferability of such systems to new settings.
Objectives
The aims of this study were to develop an AE recognition system, prospectively evaluate its performance on an external benchmark dataset and identify potential factors influencing the transferability of AE recognition systems.
Methods
A pipeline based on dictionary lookups and logistic regression classifiers was developed using a proprietary dataset of 196,533 Tweets manually annotated for AE relations and prospectively evaluated the system on the publicly available WEB-RADR reference dataset, exploring different aspects affecting transferability.
Results
Our system achieved 0.53 precision, 0.52 recall and 0.52 F1-score on the development test set; however, when applied to the WEB-RADR reference dataset, system performance dropped to 0.38 precision, 0.20 recall and 0.26 F1-score. Similarly, a previously published method aiming at automatically detecting adverse event posts reported 0.5 precision, 0.92 recall and 0.65 F1-score on thus another dataset, while performance on the WEB-RADR reference dataset was reduced to 0.37 precision, 0.63 recall and 0.46 F1-score. We identified four potential factors leading to poor transferability: overfitting, selection bias, label bias and prevalence.
Conclusion
We warn the community about a potentially large discrepancy between the expected performance of automated AE recognition systems based on published results and the actual observed performance on independent data. This study highlights the difficulty of implementing an all-purpose system for automatic adverse event recognition in Twitter, which could explain the lack of such systems in practical pharmacovigilance settings. Our recommendation is to use benchmark independent datasets, such as the WEB-RADR reference, to investigate the transferability of the adverse event recognition systems and ultimately enforce rigorous comparisons across studies on the task.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Transferability of adverse event (AE) recognition systems developed for social media has not been properly investigated so far. |
An AE recognition system for Twitter data has been developed in the course of the WEB-RADR project. The developed system and another published method for AE-post classification were prospectively evaluated on an external, independently annotated dataset and both showed a substantial drop in performance compared with reported results on the datasets used for their development. |
Relying on traditional cross-validation schemes might lead to an overestimation of the transferability of AE recognition systems in social media. This study identifies four potential factors leading to poor transferability: overfitting, selection bias, label bias and prevalence. Utilization of a benchmark independent dataset will help the community to get a better understanding of AE recognition systems on previously unseen data. |
1 Introduction
The internet has radically changed the way patients inform themselves about diseases and medicinal products [1, 2]. In a survey by the Pew Research Center, it was estimated that 6% of internet users posted comments or stories regarding personal health experiences online over 1 year, and the majority of this group did so in order to reach a general audience of friends or other internet users [3]. Twitter, a social networking service, is one of the largest social media platforms with more than 120 million daily users at the beginning of 2019. In Twitter, users post messages (with a maximum length of 140 characters at the beginning of this study, but 280 characters since 2017), that will be visible for anyone following the sender. With its massive number of users openly sharing their thoughts and experiences, Twitter has the potential to be a useful resource for post-marketing surveillance of medicines, complementing traditional pharmacovigilance tools with its unsolicited nature, timeliness and breadth of patient coverage [4].
In the last 10 years, a sizeable number of systems for automatic recognition of adverse events (AEs) in social media (including Twitter) have been published, with large variations on the actual task, from finding posts containing AEs [e.g. 5–8] to finding the location of the AE mentions within the post [e.g. 9–13], from simple extraction of the AE verbatims [e.g. 6, 14, 15] to mapping of the AE verbatims to specific terminologies [e.g. 10, 12, 16–18], from implicit attribution of the detected AEs to the drug of interest mentioned in the post [e.g. 18, 19] to classification of the relationship between drugs and AEs found [e.g. 20–23]. Therefore, when adding the heterogeneity of the datasets used (e.g. size, prevalence of AEs, number of drugs studied, number of AE types in focus), it becomes a real challenge to compare performances across studies and even assess whether the systems described are likely to perform well on previously unseen independent data [24, 25]. A recent comprehensive review of published work on the task of AE recognition in social media clearly highlights all these challenges, as well as a great number of limitations found in studies published in the field [25]. Despite the claims on the usefulness of social media data for pharmacovigilance purposes from many of the studies on the topic [e.g. 5, 21, 26–29], social media today rarely seem to be used in practical settings for all-purpose pharmacovigilance and have yet to demonstrate impact on the field [25]. A recent study by Caster et al. demonstrated the poor value of disproportionality analysis of Twitter and Facebook data for detection of new safety signals [30]. This obviously poses questions on the reason behind this reality: is the lack of implemented solutions a mere sign of the infancy of the research done, or could it be explained by the complexity of the task and the poor transferability of the developed algorithms to new data?
As pointed out earlier, ‘adverse event recognition’ can represent different underlying tasks, hence it is important to clarify the task our system is addressing, so as to avoid invalid cross-study comparisons of performance. Our system aims to automatically extract data that would be directly given as input for signal detection downstream, as done using spontaneous reporting databases, where suspected drugs and AEs are extracted from case reports and used to calculate a measure of disproportionality, facilitating identification of drug/AE pairs for further manual assessment [31]. Therefore, the task requires the identification of any medicinal product and any medical event within a given Tweet, the mapping of both types of concepts to dedicated terminologies (in our case WHODrug Global, the most comprehensive and actively used drug reference dictionary in the world, and the Medical Dictionary for Regulatory Activities, MedDRA®, the international medical terminology developed under the auspices of the International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use) and finally the characterization of the relationships between identified medical events and identified medicinal products as AE relations or not [32]. We have no requirement to find the exact locations of the products or the events within the Tweets, as is done in some other studies [9, 10, 33, 34]. Our success is measured by our ability to discover product/event combinations that represent AE relationships and appropriately map the product and the event to the correct respective entry in the terminologies. It is noteworthy to highlight the importance for downstream analysis of the mapping step so that mentions like “can’t sleep” and “still awake” can be mapped to the single concept of Insomnia. For the events, we relax the evaluation constraints by mapping the annotations produced by the system to the gold standard annotations at the Higher Level Terms (HLTs) of the MedDRA® hierarchy, to partly circumvent the potential subjectivity introduced by the manual mapping of the event terms. For precision computations, a true positive medical event is a preferred term (PT) found by the system for which at least one PT in the gold standard annotated events is found under the same HLT. Similarly, for recall computations, a true positive medical event is a PT from the gold standard annotations for which at least one PT annotation has been found by the system under the same HLT. The two kinds of true positives might not match exactly.
Only a limited number of published systems aim at accomplishing this comprehensive task, most other systems are designed to target very specific products or events or solve partial aspects of the task and would therefore need additional steps if used in routine pharmacovigilance methods such as disproportionality analysis. This reality might be explained by the scarcity of available data to train algorithms to perform the comprehensive task. The 2017 shared task from the Social Media Mining for Health (SMM4H) workshop [11] provided a valuable opportunity to compare performance of different systems (13 different teams participated) but was divided into the following subtasks: (1) binary classification of AE posts, (2) medication intake classification and (3) mapping of AE expressions to MedDRA®. However, the shared task was renewed in September 2019 [35] and included one task aiming to jointly find the mentions of AEs and map the expressions to MedDRA®. Although the task did not include recognition of the drug, the best system obtained an F-score of 0.432 [35], which is much lower than most published results of so-called ‘AE recognition systems’ [25]. Among other publicly available datasets, we found two others that were compatible with the development of a system that solves the comprehensive AE recognition task (i.e. find all drug mentions, all event mentions, map them to respective terminologies and finally characterize their relationships): the CADEC corpus [36] and the TwiMed corpus [37]. Nonetheless, the vast majority of systems developed using these two datasets focused on one single subtask, the location of the ADR mention for systems using the CADEC corpus [e.g. 13, 38–40] and post-/sentence-level AE classification for the TwiMed corpus [e.g. 41], with the mapping of the event mention to a terminology being ignored. Solving the comprehensive task within one single dataset has proven to be challenging [35]. Therefore, serious concerns about the ability of AE recognition systems to maintain their performance in applied settings (typically, new streams of social media data) can be raised, as such transfer is likely to cause a certain degree of performance drop. However, the question of transferability of such systems to new data has been largely left unaddressed.
With this study, we provide a first attempt at answering this question. The study is embedded in a larger project, carried out by the WEB-RADR consortium, a partnership between academia, industry and regulators and supported by the Innovative Medicines Initiative Joint Undertaking. One of the goals of the WEB-RADR project was to investigate the usefulness of social media for pharmacovigilance [42]. The issue raised by the question above highlights the great need for benchmark datasets, used solely for evaluation purposes. Because annotated datasets are scarce, such datasets are often used both for training and evaluation. This means that the transferability of most developed systems, that is, the ability to maintain acceptable performance when applied in new contexts, is basically unknown. In fact, out of all the studies (~ 50) we have compiled in relation to the topic, none provide any sort of external validation for their developed systems of AE recognition. Poor transferability is likely to affect the more sophisticated methods, trained on datasets of limited size. To our knowledge, this study is the first to present the development of an AE recognition system together with a prospective evaluation of its performance outside of the universe of the data it has been trained on. We perform an external evaluation using a publicly available benchmark dataset manually curated and annotated by members of the WEB-RADR consortium [43]. The dataset is entirely independent from the dataset we used for training our system, which was provided to us by Epidemico, a health informatics company (later acquired by Booz Allen Hamilton) and former WEB-RADR partner. Epidemico also published on a system for the recognition of posts with AE mentions as well as their characterization [5, 18], which seems to achieve state-of-the-art performance on the task. In this paper, we present external evaluation results for both our system and the system described in [18]. In addition, we sought to provide preliminary answers regarding the observed performance difference when the systems are applied to their respective training datasets and when they are applied to previously unseen and independently annotated data. Another noteworthy original aspect of the system developed in this work is its scope: by design, it aims at finding any type of AE for any kind of medicinal product, a necessary requirement for performing pharmacovigilance on a global scale.
2 Methods
2.1 System Overview
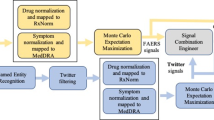
The automatic recognition and mapping of AEs in Twitter posts developed in this study is implemented like a pipeline, where Tweets flow through different components (modules) aimed at solving specific subtasks before finally being converted into a list of medicinal product/medical event pairs with a suspected AE relationship between them. There are three modules in our system. First, a relevance filter discards Tweets with low resemblance to AE posts, using the Indicator Score introduced elsewhere [18]. Second, a Named Entity Recognition (NER) module recognizes mentions of products as well as events, and then maps the recognized mentions to standardized terminologies (WHODrug and MedDRA® PTs, respectively). Finally, an AE relation classification module classifies all possible pairs of recognized products and events as AE relations or not (Fig. 1). To provide a good trade-off between readability and reproducibility, a brief description of the datasets and the methods involved in the three modules is given in the following subsections, however, the more technical details are provided in Online Resource 1 (see electronic supplementary material [ESM]).
Although we have arranged the system so that the relevance filter comes before the NER and mapping module, both modules are independent and thus could be applied in the reverse order. In the result section, we thus provide a detailed view of how AE relations are lost in these two modules, ignoring the order in which they are applied. This allows us to assess the performance of both modules separately.
2.2 Datasets
The system partly involves machine learning methods. To train the associated models, we have used a proprietary dataset provided by Epidemico, of 196,533 manually annotated Tweets (see [5] for a description of the annotation process), which after de-duplication and pre-processing (e.g. English language filtering) resulted in 138,885 Tweets further divided into a training set to learn the parameters of all models, a validation set to tune the hyperparameters and a test set for evaluation of the system (97,190/27,963/13,732 Tweets, respectively). The entire processed proprietary dataset involves 125,660 medicinal product annotations (862 unique products) and 92,909 medical event annotations (507 unique MedDRA® PTs), for a total of 37,434 AE relations (25,125/8762/3547 for the training, validation and test sets, respectively), representing one of the largest AE-annotated Twitter training datasets to date. We refer to this dataset as the system dataset, to keep in mind the tight relation existing between the dataset and the system, as all parameters of the system are trained using this dataset.
A second dataset is used in this study, to provide an external prospective validation of the system and to provide an idea of its transferability: a publicly available set of 57,473 Tweets manually curated for AE relations, developed in the course of the WEB-RADR project and intended as a benchmark for the task [43]. In this dataset, only Tweets with valid AE relations are annotated for medicinal products of interest and medical events, as well as the AE relations. There are 1056 Tweets with at least one AE relation (AE posts) and 1396 AE relations in total. We refer to this dataset as the reference dataset.
2.3 Relevance Filter
To increase the proportion of relevant posts, we apply a previously published method to score every post for their resemblance to posts containing AE relations [5, 18, 44]. In brief, each Tweet is converted into a bag of words. Under a Bayesian probabilistic model, a composite score—called the Indicator Score—is calculated based on the likelihood that the Tweet contains an AE combined with the likelihood that the Tweet does not contain an AE. An Indicator Score can lie between 0 and 1, with values close to 1 suggesting the presence of at least one AE mention in the post. Posts with scores above 0.7 were retained while the others were discarded, as was done in [18].
2.4 Named Entity Recognition and Mapping
Product names are recognized via dictionary lookup using WHODrug Global (Uppsala Monitoring Centre, Uppsala). Dictionary entries with a high level of ambiguity (such as the tradename ‘Today’) are removed automatically before the lookup, to reduce noise. Overlapping matches are resolved by match size [45]. As we are using WHODrug Global, mapping to substances is trivial.
Medical events are recognized via dictionary lookups and machine learning. The first dictionary used is MedDRA® Lowest Level Terms. The second dictionary is extracted from VigiBase, the World Health Organization (WHO) global database of individual case safety reports. By using the reported verbatim descriptions of reactions, we include more expressions related to medical events. Finally, we train 169 logistic regressions using the system dataset. Each logistic regression uses the Tweet as a bag-of-grams (up to tri-grams) as input and targets a single MedDRA® Preferred Term that has been annotated at least 20 times in the training dataset. We only retained the 124 logistic regression models for which the validation performance exceeded 0.4 in F-score. Mapping of the events is thus done to MedDRA® PTs directly by design.
2.5 Adverse Event Relation Classifier
After the NER and mapping module, every possible pair of a medicinal product and a medical event that have been recognized in a Tweet that satisfied the Indicator Score threshold is evaluated for their AE relation. We trained a logistic regression classifier based on document features (e.g. number of URLs, of words, of user mentions), on syntactic features (e.g. product before event, number of words between the product mention and the event mention) and semantic features using word2vec [46] representations clustered in discrete groups. The full list of features used in the model is given in the Online Resource 1 (see ESM). Word2vec is an algorithm that can automatically, and without supervision, learn vector representations of words using a very large amount of text. Words appearing in similar contexts end up with similar vector representations, leading to a (usually) high-dimensional space of meaning, where neighbouring words have similar meaning. Word vectors can provide a level of abstraction that go beyond the mere terms employed. We used existing word vectors pre-trained on a large corpus of 400 million Tweets [47]. However, we did not use the vectors directly, instead we clustered the word vectors (500 different clusters, using K-means clustering), as has been successfully done in a previous study [9].
3 Results
3.1 Performance Results
The recall performance results of the first two components, the relevance filter and the NER module, are summarized in Fig. 2 as a Venn diagram. As can be seen in the intersection of the three module parts, only 68.4% of the 25,125 AE relations of the training set are still discoverable after the first two components (appearing in a post of Indicator Score > 0.7 and having both product and event correctly recognized by the NER module), before applying the AE relation classifier. This number drops moderately to 63.3% for the test dataset and considerably to 30.4% for the WEB-RADR reference dataset. This means that 31.6% of the AE relations in the training set are being lost by either appearing in a Tweet with Indicator Score < 0.7, or by having its medicinal product not recognized, or by having its medical event not recognized, while this percentage increases to 36.7% for the test set and to 69.6% for the WEB-RADR reference dataset. For all three datasets, the event recognition component is the main bottleneck, with 17.3%, 21.0% and 26.8% of the AE relations passing the relevance filter and having their product correctly recognized but their event either not detected or improperly mapped to MedDRA® in the training, test and WEB-RADR reference dataset, respectively (see the intersection between the green and blue ovals in Fig. 2).
Performance in recalling adverse event (AE) relations of the relevance filter and the Named Entity Recognition (NER) and mapping module. The total number of AE relations of the training set, the test set and the WEB-RADR reference set is given on the upper right corner. The figures in the Venn diagram indicate the percentage of AE relations correctly passing or failing the different module parts
The product NER is the only component that does not display a drop in performance when evaluating the WEB-RADR data, with 0.878 recall of the products in AE relations in the test data versus 0.896 in the WEB-RADR reference data (this can be computed by summing all percentages in the blue oval in Fig. 2, representing all AE relations for which the medicinal product gets correctly recognized). It should be noted that WHODrug, used in this system, has also been used in the development of the WEB-RADR reference dataset to provide search terms for the six substances of interest; hence, the recall is expected to be smaller when considering all possible product mentions that could exist in the world. In contrast, the event NER displays a drop in recall from 0.743 of the events involved in AE relations in the test dataset to 0.461 in the reference dataset, and the relevance filter as well, from 0.953 of AE relations passing the filter in the test set to 0.644 in the reference dataset (this can be computed by summing all percentages in the pink oval in Fig. 2, representing all AE relations for which the medical event gets correctly recognized). The absolute drop in recall of the relevance filter and both NER modules between the training dataset and the test dataset is much more moderate (0.01 for the relevance filter, 0.009 for the product NER and 0.055 for the event NER).
Considering the detection of AE posts (as opposed to AE relations), the Indicator Score gave a precision of 0.63 and recall of 0.96 (F1-score 0.76) on the test dataset, which exceeds published performance (0.50 precision, 0.92 recall and 0.65 F1-score [18]). As the system dataset might include posts used to train the Indicator Score, this performance result is likely to represent an overestimation of the performance that can be expected on new datasets. In fact, we also observed a clear drop in performance of the Indicator Score when applying to the reference dataset (0.37 precision, 0.63 recall and 0.46 F1-score).
Out of the 17,175 true AE relations still retained in the training set after the relevance filter and the NER module (i.e. 68.4% of the original 25,125 AE relations), 14,608 were correctly classified as AE relations by the AE relations classifier, which represents a recall of 0.85 for the classifier alone and a recall of 0.58 for the entire system. For the test set, the recall of the classifier drops to 0.80 and the overall recall to 0.52. For the WEB-RADR reference dataset, the recall of the classifier is 0.63 and the overall recall is 0.20.
Precision-wise, the NER module produces many potential product/event combinations to be classified as AE relations or not (Table 1). In all three datasets, the AE relation classifier manages to improve the precision of the product/event combinations obtained after the NER module, from 0.31 pre-classification to 0.61 post-classification in the training set, 0.28 to 0.53 in the test set and 0.27 to 0.38 in the WEB-RADR dataset; however, the benefit is much more marginal in the reference dataset compared with the other two datasets.
Overall, the system obtains the following performance results for recognizing, correctly coding and correctly classifying AE relations: 0.61 precision, 0.58 recall and 0.60 F1-score on the training set, 0.53 precision, 0.52 recall and 0.52 F1-score on the test set, and finally 0.38 precision, 0.20 recall and 0.26 F1-score on the independently annotated WEB-RADR reference dataset. The F1-score of the entire AE recognition system is thus halved when moving from the test set to the independent WEB-RADR reference data.
3.2 One Size Does Not Fit All
There are 291 unique PTs annotated in the WEB-RADR reference data, and the majority of them (156) are annotated only once in the dataset. When comparing F1-score performance broken down by PT between the test set and the reference dataset, we observe that the vast majority of PTs have a lower observed performance in the reference dataset (see Fig. 3). The performance of our system on the ten most commonly annotated PTs in the reference dataset is summarized in Table 2.
The use of dictionary lookups in the event NER allows the system to identify medical events that have never been observed in the training data. However, the performance is limited by the richness of expressions related to the medical events, which can only be captured if the dictionaries contain those expressions (e.g. ‘drug use disorder’ in Table 2). The PT ‘social problem’ illustrates another important limitation of recognizing medical events: the subjectivity of the annotation. Most Tweets annotated for this PT describe the discontentment of the author to one associated product (e.g. “[…] Drug X is the most horrific drug.[…]”, “Fucking Drug X and Drug Y h8 u both”, “Talking about Drug X makes me sad”). Detecting common patterns in those Tweets is challenging for the algorithm. In fact, even describing these as medical events and characterizing them as AE relations to the product can be seen as debatable.
3.3 Error Analysis
Out of the 1396 AE relations present in the WEB-RADR reference dataset, 1114 of them have been missed by the system. There are four possible sources of false negatives: the system can have missed the product mention or miscoded it (146 AE relations), the system can have missed the event mention or miscoded it (753 AE relations), the AE relation can be in a post that did not pass the relevance filter (498 AE relations) or in a post with Indicator Score > 0.7 with both product and event correctly coded, but mistakenly classified as a non-AE relation (210 AE relations). Note that the first three sources of false negatives just mentioned are not mutually exclusive. The event NER thus represents the major bottleneck in recall for the reference dataset, followed by the Indicator Score filter.
We also analysed the 482 product–event pairs that the system mistakenly classified as AE relations. Of these, 52 (11%) could potentially be interpreted as AE relations, and 77 (16%) could be associated with different annotation practices used between the system dataset and the reference dataset. The major source of these differences was related to expressions of psychotic effects such as ‘high’, ‘doped up’, ‘floating around’ or ‘loopy’. These were generally mapped to the PT Altered state of consciousness in the system dataset (the machine learning component of the event recognition thus learned to make these associations), while they were mapped to Euphoric mood or Feeling abnormal in the reference dataset. Another example of these coding differences relates to the coding of unspecific expressions such as ‘side effects’, which were mapped to Nonspecific reaction in the system dataset while mapped to Adverse drug reaction or Adverse event in the reference dataset. These types of errors are problematic for a truthful evaluation of the system, because they actually lead to two paired errors: a false positive error where the event is coded according to the annotation practices of the training dataset, and a false negative error where the event is coded according the annotation practices of the independent dataset. Evaluation of the system at the HLT level instead of the PT level can mitigate these kinds of errors only to a certain degree.
Among the remaining 352 false positive AE relations that truly were mistakes of the system, a majority (141) were due to the recognition of an event unrelated to the meaning of the post. Nonetheless, we also found examples of missed negations (the event is in the text and properly coded but the author means the event did not happen), events paired with another product mentioned in the post, not with the product of interest, events that were not AEs in this context (oftentimes indication), or events related to the gold standard annotation but too general (e.g. Pain vs Injection site pain) or slightly off compared with the gold standard annotation (e.g. ‘sleepy’ often got coded by the system as Tiredness instead of Somnolence).
4 Discussion
In this study, we developed a system to automatically recognize medicinal products and medical events in Tweets, map them to WHODrug Global and MedDRA® PT terminologies, and classify product/event pairs as representing AE relations or not. The obtained performance of the system on the training dataset was 0.61 precision, 0.58 recall and 0.60 F1-score. The typical approach for estimating the future performance of systems of our kind is by means of retrospective analysis. A separate test set is reserved from the available data and used for computing measures of performance. When evaluating our system using this approach, we obtained a moderate drop in performance: 0.53 precision, 0.52 recall and 0.52 F1-score.
Measures obtained this way can, however, be expected to be biased, because product and AE mapping conventions, the set of monitored products and safety profiles, epidemiological aspects of the population at the site of implementation, and the prevalence of reported AEs, may vary. A more realistic estimate of future performance can be obtained by instead performing a prospective evaluation of the system on data collected after completion of the system, from the context and under conditions where the system will be implemented. The present study is to our knowledge the first attempt to prospectively evaluate the performance of an AE recognition system for social media. Our evaluation, using an external, independently annotated dataset, resulted in a significant drop in performance compared with our retrospective evaluation: 0.38 precision (two-third of the training precision), 0.20 recall (one-third of the training recall) and 0.26 F1-score (less than half of the training F1-score).
None of the published studies that we reviewed had, however, performed such an evaluation, despite its potential for revealing positive biases in estimated performances. Comparing AE recognition performance of our system with other published systems is problematic not only because of different study designs, but also because most published studies addressed fewer or different tasks. If we were to ignore mapping, focus on the task of identifying AE posts and classify any post with a relation classified as AE by our system, we would obtain 0.76 precision, 0.67 recall and 0.71 F1-score for detecting AE posts when applied to the test dataset, which is in the range of published results. Detecting AE posts this way leads to 0.70 precision, 0.39 recall and 0.50 F1-score when applied to the reference dataset, clearly better performance results than the results presented in the above paragraph (in fact, F1-score on the reference dataset is doubled for the AE-post recognition task compared with the full AE recognition task).
Another method that similarly presented poor transferability when evaluated on the WEB-RADR reference dataset is the Indicator Score method, which aims to detect posts with high resemblance to AE posts [18]. In the study, Powell and colleagues found that using an Indicator Score threshold of 0.7 led to a precision of 0.50 and a recall of 0.92 for finding AE posts (0.65 F1-score). On the WEB-RADR reference dataset, this performance dropped to 0.37 precision and 0.63 recall (0.46 F1-score). While the drop in precision could potentially be explained by the different prevalence of AE posts in the dataset used in the publication and in the reference dataset (25% vs 1.8%), recall is not expected to depend on prevalence and thus there must be other explanations for its performance drop. AE recognition is not the first natural language processing task to have poor transferability when applied to external datasets. Negation detection algorithms have also demonstrated similar difficulties [48].
The use of an external independent annotated dataset (the WEB-RADR reference) gave us a unique opportunity to study the effects of transferability of AE recognition systems. We warn the community on the existence of several potential factors that can lead to poor transferability. One factor that can affect machine-learning–based methods is overfitting. It is illustrated by the drop in performance observed for the Indicator Score filter as well as for the AE classifier module and for the event recognition component of the NER module. The latter provides the most compelling illustration. The event recognition component has two parts: a dictionary lookup part based on MedDRA® lowest level terms and VigiBase reported reactions, and a machine-learning-based part composed of 124 logistic regressions. The dictionary lookup part was unaffected by the transfer to a new dataset (0.35 recall of the events involved in AE relations of the test set vs 0.33 recall in the reference dataset). In contrast, the machine-learning-based part was clearly affected (0.68 vs 0.32 recall, respectively). This overfitting actually happens at the level of the entire training dataset (the system dataset in our case), not just on the training part of the traditional training/validation/test split. The machine-learning–based part of the event recognition component had indeed a much more moderate drop in performance when comparing performance on the training set (0.75 recall) to the performance on the test set (0.68 recall). The dataset-level overfitting is tightly linked to our second identified factor for explaining poor transferability of AE recognition systems: the issue of systematic differences between the training dataset and the external dataset.
Systematic differences between datasets cannot be alleviated by the typical methods of overfitting reduction (e.g. cross-validation, regularization). We have identified two main sources of those differences: selection bias and label bias. Selection bias relates to all factors that contributed to making the datasets, selecting the Tweets. In most AE recognition studies, Tweets are collected via Twitter API using search terms that often represent tradenames and substances of interest. Differing products leads to different safety profiles (the AEs will be different in nature) and different kinds of users (e.g. age, sex), which, combined, can lead to very different ways of expression. For instance, methylphenidate users are likely to differ from interferon users; they will tend to express themselves differently in their posts, and the kind of events they will talk about will also differ. The WEB-RADR reference dataset has only six substances of interest: Methylphenidate (34.2%, used to treat attention-deficit disorders), Zolpidem (30.3%, used to treat insomnia), Levetiracetam (23.8%, an anti-epileptic), Insulin glargine (6.8%, used to treat diabetes), Terbinafine (3.5%, an anti-fungal drug) and Sorafenib (1.3%, used to treat advanced renal cell carcinoma). Although these substances do appear in the system dataset, they only represent 5.6% of the product mentions in AE relations. In contrast, the top six substances associated with AE relations in the system dataset are Ibuprofen (11.7%, a non-steroidal anti-inflammatory drug), Alprazolam (3.5%, an anxiolytic), Paracetamol (3.3%), Human papilloma virus vaccine (3.1%), Zolpidem (3.0%) and Oxycodone (3.0%, an opioid used to treat severe pain). Apart from the Tweets involving zolpidem, the expressions found in the two datasets are likely to differ, because the products being discussed are very different.
The second source of systematic differences between datasets is label bias. Label bias relates to the subjectivity surrounding the annotation of the datasets. Tweets are short and often quite informal. It can sometimes be hard to interpret what the author means. In the case of classifying a Tweet as containing the mention of an adverse drug reaction or not, different annotators can reach different conclusions. In a study based on Twitter data, a Kappa value (inter-annotator agreement) of 0.69 has been found for this task [49], which demonstrates a non-negligible level of subjectivity. Most annotation work involves initial cycles of annotations where annotators develop guidelines in order to achieve a high consensus in their annotations. Interpretations regarding what counts as an AE might differ, and so can the mapping of the associated event. In the error analysis, 16% of the false positives could be attributed to different practices in the coding of the event between the system dataset and the reference dataset. This kind of bias is only problematic if the system data is gathered from an external source and applied to another dataset of interest. Annotation practices adopted for making the training set of the AE recognition system will impact the results obtained on new data. If there are systematic differences in how the annotation is desired versus how it is produced by the system, some additional automatic corrective steps can be taken (e.g. mapping Altered state of consciousness to Feeling abnormal under some conditions related to the Tweet text). However, if there are no clear rules that can be derived to achieve the desired annotation practice, the system might have to be re-trained in-house, with a dataset whose annotations are following the desired practices.
Finally, another factor that can affect performance results across datasets, especially precision results, is prevalence. Regrettably, few studies clearly specify the prevalence of AEs in their training data or discuss the implications of that prevalence on their results as well as the transferability of their performance results to more real-world settings. In most studies, the annotated training dataset is enriched with AE mentions compared with what we expect to find in Twitter. When applied to low AE prevalence data, algorithms trained on high AE prevalence data are likely to display a dramatic decrease in precision (and thus in F1-score to a smaller extent). In social media such as Twitter, where a tiny proportion of posts about medicinal products is expected to contain AE mentions, this effect is likely to be exacerbated.
5 Conclusion
There is a great need for external evaluation of AE recognition systems developed for Twitter, and probably social media in general. The field seems to suffer from a lack of reproducibility. Although efforts have been made for ensuring fair comparison between systems [11, 35], additional publicly available annotated benchmark datasets, used solely for evaluation purposes, could help the field progress and allow for more comparisons across studies, notably on their ability to generalize to new data. In this study, by using the WEB-RADR reference dataset, a publicly available dataset [43], we identified a number of factors that could explain the poor transferability of the system we developed and of another published system aimed at classifying AE posts. The poor transferability offers a plausible explanation to why, despite almost a decade since the first AE recognition systems in social media have been published, such systems have not been adopted in routine pharmacovigilance practice. The vision of an all-purpose social-media-based pharmacovigilance system can only be attained if a reliable and performant AE recognition system is developed. Another study performed under the umbrella of the WEB-RADR project used a state-of-the-art AE recognition system to identify AE relations from Twitter and Facebook posts and applied statistical signal detection methods [30]. Caster and colleagues found that these social media had no predictive value for either labelling changes or validated signals, and they point at the AE recognition system as one of the limiting factors that could explain their results. Such a finding puts the use of social media for pharmacovigilance into serious question. As a community, we may have to re-think how social media could be of use for detecting safety concerns in the use of medicines. It might be that an all-purpose (all products, all events) pharmacovigilance system is unfeasible, but questions of more limited scope (e.g. studies of lack of effect or drug abuse) could still be addressed using this kind of data. Mining dedicated forums could provide data of higher quality and help investigate targeted issues. In any case, it seems clear that the utility of social media for pharmacovigilance remains an open question and that additional, carefully described research is needed to really understand the value social media could represent for monitoring the safety of medicinal products.
References
Harpaz R, Callahan A, Tamang S, Low Y, Odgers D, Finlayson S, Jung K, LePendu P, Shah NH. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug Saf. 2014;37(10):777–90.
Moorhead SA, Hazlett DE, Harrison L, Carroll JK, Irwin A, Hoving C. A new dimension of health care: systematic review of the uses, benefits, and limitations of social media for health communication. J Med Internet Res. 2013;15(4):e85.
Pew Research Center: Internet ST. Health Online 2013 [Internet]. Pew Research Center; 2015. https://www.pewinternet.org/2013/01/15/health-online-2013/. Accessed 6 Sept 2018.
Sarker A, Ginn R, Nikfarjam A, O’Connor K, Smith K, Jayaraman S, Upadhaya T, Gonzalez G. Utilizing social media data for pharmacovigilance: a review. J Biomed Inform. 2015;1(54):202–12.
Freifeld CC, Brownstein JS, Menone CM, Bao W, Filice R, Kass-Hout T, Dasgupta N. Digital drug safety surveillance: monitoring pharmaceutical products in twitter. Drug Saf. 2014;37(5):343–50.
Alvaro N, Conway M, Doan S, Lofi C, Overington J, Collier N. Crowdsourcing Twitter annotations to identify first-hand experiences of prescription drug use. J Biomed Inform. 2015;58:280–7.
Patki A, Sarker A, Pimpalkhute P, Nikfarjam A, Ginn R, O’Connor K, et al. Mining adverse drug reaction signals from social media: going beyond extraction. Proc BioLinkSig. 2014;2014:1–8.
Rastegar-Mojarad M, Elayavilli RK, Yu Y, Liu H. Detecting signals in noisy data-can ensemble classifiers help identify adverse drug reaction in tweets. In: Proceedings of the Social Media Mining Shared Task Workshop at the Pacific Symposium on Biocomputing 2016.
Nikfarjam A, Sarker A, O’connor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Inform Assoc. 2015;22(3):671–81.
Karimi S, Metke-Jimenez A, Nguyen A. CADEminer: a system for mining consumer reports on adverse drug side effects. In: Proceedings of the eighth workshop on exploiting semantic annotations in information retrieval: 2015: ACM, 2015; p. 47–50.
Sarker A, Belousov M, Friedrichs J, Hakala K, Kiritchenko S, Mehryary F, Han S, Tran T, Rios A, Kavuluru R, de Bruijn B. Data and systems for medication-related text classification and concept normalization from Twitter: insights from the Social Media Mining for Health (SMM4H)-2017 shared task. J Am Med Inform Assoc. 2018;25(10):1274–83.
Metke-Jimenez A, Karimi S. Concept extraction to identify adverse drug reactions in medical forums: a comparison of algorithms. arXiv preprint arXiv:150406936. 2015.
Tutubalina E, Nikolenko S. Combination of deep recurrent neural networks and conditional random fields for extracting adverse drug reactions from user reviews. J Healthc Eng. 2017;2017:1–9.
Liu J, Li A, Seneff S. Automatic drug side effect discovery from online patient-submitted reviews: focus on statin drugs. In: Proceedings of First international conference on advances in information mining and management (IMMM): Barcelona, Spain; 2011. p. 23–9.
Risson V, Saini D, Bonzani I, Huisman A, Olson M. Validation of social media analysis for outcomes research: identification of drivers of switches between oral and injectable therapies for multiple sclerosis. Value Health. 2015;18(7):A729.
Chee BW, Berlin R, Schatz B. Predicting adverse drug events from personal health messages. AMIA Annu Symp Proc. 2011;2011:217–26.
Leaman R, Wojtulewicz L, Sullivan R, Skariah A, Yang J, Gonzalez G. Towards internet-age pharmacovigilance: extracting adverse drug reactions from user posts to health-related social networks. In: Proceedings of the 2010 Workshop on Biomedical Natural Language Processing. USA: Association for Computational Linguistics; 2010. p. 117–25.
Powell GE, Seifert HA, Reblin T, Burstein PJ, Blowers J, Menius JA, et al. Social media listening for routine post-marketing safety surveillance. Drug Saf. 2016;39(5):443–54.
Freifeld CC. Digital Pharmacovigilance: the medwatcher system for monitoring adverse events through automated processing of internet social media and crowdsourcing. 2014. Doctoral dissertation, Boston University.
Nikfarjam A, Gonzalez GH. Pattern mining for extraction of mentions of Adverse Drug Reactions from user comments. AMIA Annu Symp Proc. 2011;2011:1019–26.
White RW, Harpaz R, Shah NH, DuMouchel W, Horvitz E. Toward enhanced pharmacovigilance using patient-generated data on the internet. Clin Pharmacol Ther. 2014;96(2):239.
Yang CC, Yang H, Jiang L, Zhang M. Social media mining for drug safety signal detection. Proceedings of the 2012 international workshop on smart health and wellbeing - SHB ’12 [Internet]. Maui, Hawaii, USA: ACM Press; 2012 p. 33. Available from: http://dl.acm.org/citation.cfm?doid=2389707.2389714.
Sampathkumar H, Chen X-W, Luo B. Mining adverse drug reactions from online healthcare forums using hidden Markov model. BMC Med Inform Decis Mak. 2014;14(1):1.
Chen X, Deldossi M, Aboukhamis R, Faviez C, Dahamna B, Karapetiantz P, et al. Mining adverse drug reactions in social media with named entity recognition and semantic methods. Stud Health Technol Inform. 2017;245:322–6.
Tricco AC, Zarin W, Lillie E, Jeblee S, Warren R, Khan PA, Robson R, Hirst G, Straus SE. Utility of social media and crowd-intelligence data for pharmacovigilance: a scoping review. BMC Med Inform Decis Mak. 2018;18(1):38.
Topaz M, Lai K, Dhopeshwarkar N, Seger DL, Sa’adon R, Goss F, et al. Clinicians’ reports in electronic health records versus patients’ concerns in social media: a pilot study of adverse drug reactions of aspirin and atorvastatin. Drug Saf. 2016;39(3):241–50.
Akay A, Dragomir A, Erlandsson B-E. Network-based modeling and intelligent data mining of social media for improving care. IEEE J Biomed Health Inform. 2015;19(1):210–8.
Dole O. Discovering drug side effects with crowdsourcing. 2013. https://www.crowdflower.com/discovering-drug-side-effects-with-crowedsourcing/. Accessed June 2019.
Pages A, Bondon-Guitton E, Montastruc JL, Bagheri H. Undesirable effects related to oral antineoplastic drugs: comparison between patients’ internet narratives and a national pharmacovigilance database. Drug Saf. 2014;37(8):629–37.
Caster O, Dietrich J, Kürzinger ML, Lerch M, Maskell S, Norén GN, Tcherny-Lessenot S, Vroman B, Wisniewski A, van Stekelenborg J. Assessment of the utility of social media for broad-ranging statistical signal detection in pharmacovigilance: results from the WEB-RADR project. Drug Saf. 2018;41(12):1355–69.
Meyboom RH, Egberts AC, Edwards IR, Hekster YA, de Koning FH, Gribnau FW. Principles of signal detection in pharmacovigilance. Drug Saf. 1997;16(6):355–65.
Pappa D, Stergioulas LK. Harnessing social media data for pharmacovigilance: a review of current state of the art, challenges and future directions. Int J Data Sci Anal. 2019;8:1–23.
Sarker A, Nikfarjam A, Gonzalez G. Social media mining shared task workshop. Biocomputing 2016. Kohala Coast, Hawaii, USA: World Scientific; 2016. p. 581–92. Available from: http://www.worldscientific.com/doi/abs/10.1142/9789814749411_0054.
Jimeno-Yepes A, MacKinlay A, Han B, Chen Q. Identifying diseases, drugs, and symptoms in twitter. Stud Health Technol Inform. 2014;216:643–7.
Weissenbacher D, Sarker A, Magge A, Daughton A, O’Connor K, Paul M, Gonzalez G. Overview of the Fourth Social Media Mining for Health (SMM4H) Shared Tasks at ACL 2019. In: Proceedings of the Fourth Social Media Mining for Health Applications (# SMM4H) Workshop & Shared Task; 2019. p. 21–30.
Karimi S, Metke-Jimenez A, Kemp M, Wang C. Cadec: a corpus of adverse drug event annotations. J Biomed Inform. 2015;1(55):73–81.
Alvaro N, Miyao Y, Collier N. TwiMed: Twitter and PubMed comparable corpus of drugs, diseases, symptoms, and their relations. JMIR Public Health Surveill. 2017;3(2):e24.
Stanovsky G, Gruhl D, Mendes P. Recognizing mentions of adverse drug reaction in social media using knowledge-infused recurrent models. Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics: Volume 1, Long Papers. 2017. p. 142–51.
Alimova I, Tutubalina E. Automated detection of adverse drug reactions from social media posts with machine learning. In: van der Aalst WMP, Ignatov DI, Khachay M, Kuznetsov SO, Lempitsky V, Lomazova IA, et al., editors. Analysis of images, social networks and texts. Cham: Springer International Publishing; 2018. p. 3–15. https://doi.org/10.1007/978-3-319-73013-4_1
Tang B, Hu J, Wang X, Chen Q. Recognizing continuous and discontinuous adverse drug reaction mentions from social media using LSTM-CRF. Wirel Commun Mob Comput. 2018;2018:1–8.
Zhang T, Lin H, Ren Y, Yang L, Xu B, Yang Z, Wang J, Zhang Y. Adverse drug reaction detection via a multihop self-attention mechanism. BMC Bioinform. 2019;20(1):479.
WEB-RADR, https://web-radr.eu. Accessed 5 Apr 2019.
Dietrich J, Gattepaille LM, Grum BA, Jiri L, Lerch M, Sartori D, Wisniewski A. Adverse events in social media—development of a gold standard reference set: results from the WEB-RADR Project. Drug Saf. 2020. https://doi.org/10.1007/s40264-020-00912-9.
Pierce CE, Bouri K, Pamer C, Proestel S, Rodriguez HW, Van Le H, Freifeld CC, Brownstein JS, Walderhaug M, Edwards IR, Dasgupta N. Evaluation of facebook and twitter monitoring to detect safety signals for medical products: an analysis of recent FDA safety alerts. Drug Saf. 2017;40(4):317–31.
Hedfors S, Bergvall T, Gilbert M, Pierce C, Dasgupta N, Ellenius J. Improving the yield of relevant data for pharmacovigilance analysis by reducing search term complexity—a study on reddit data. Abstract Pharmacoepidemiol Drug Saf. 2016;25(S3):412–3.
Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. In: Burges CJC, Bottou L, Welling M, Ghahramani Z, Weinberger KQ, editors. Advances in neural information processing systems 26. Curran Associates, Inc.; 2013. p. 3111–9. Available from: http://papers.nips.cc/paper/5021-distributed-representations-of-words-and-phrases-and-their-compositionality.pdf.
Godin F, Vandersmissen B, De Neve W, Van de Walle R (2015) Named entity recognition for Twitter microposts using distributed word representations. Workshop on Noisy User-generated Text, ACL 2015.
Wu S, Miller T, Masanz J, Coarr M, Halgrim S, Carrell D, Clark C. Negation’s not solved: generalizability versus optimizability in clinical natural language processing. PLoS One. 2014;9(11):e112774.
Ginn R, Pimpalkhute P, Nikfarjam A, Patki A, O’Connor K, Sarker A, et al. Mining Twitter for adverse drug reaction mentions: a corpus and classification benchmark. Proceedings of the fourth workshop on building and evaluating resources for health and biomedical text processing. 2014. p. 1–8.
Acknowledgements
MedDRA® trademark is registered by IFPMA on behalf of ICH. The authors are indebted to the national centres who make up the World Health Organization Programme for International Drug Monitoring and contribute reports to VigiBase. However, the opinions and conclusions of this study are not necessarily those of the various centres nor of the World Health Organization. The authors further thank Ola Caster for valuable comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
The research leading to these results was conducted as part of the WEB-RADR consortium (https://webradr.eu), which is a public–private partnership coordinated by the Medicines and Healthcare products Regulatory Agency. The WEB-RADR project has received support from the Innovative Medicine Initiative Joint Undertaking (https://www.imi.europa.eu) under Grant Agreement No. 115632, resources of which are composed of financial contributions from the European Union’s Seventh Framework Programme (FP7/2007–2013) and the European Federation of Pharmaceutical Industries and Associations companies’ in-kind contribution.
Conflict of interest
Carrie E. Pierce has no conflict of interest directly relevant to the content of this study. Lucie M. Gattepaille, Sara Hedfors Vidlin, Tomas Bergvall and Johan Ellenius are employed by the Uppsala Monitoring Centre, a non-profit foundation that commercializes WHODrug Global, the drug dictionary used in this study.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
The training data, generously provided by the WEB-RADR consortium member Epidemico Inc. (now part of Booz Allen Hamilton), is proprietary and cannot be shared with others. The WEB-RADR reference dataset used for prospective evaluation is publicly available at https://link.springer.com/article/10.1007%2Fs40264-020-00912-9.
Code availability
Code is unavailable.
Additional information
Carrie E. Pierce: Independent researcher, USA.
Portions of the work performed by Carrie E. Pierce were undertaken while employed at Booz Allen Hamilton (formerly Epidemico Inc.), 268 Newbury Street Suite 2, Boston, MA 02116, United States.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Gattepaille, L.M., Hedfors Vidlin, S., Bergvall, T. et al. Prospective Evaluation of Adverse Event Recognition Systems in Twitter: Results from the Web-RADR Project. Drug Saf 43, 797–808 (2020). https://doi.org/10.1007/s40264-020-00942-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40264-020-00942-3