Abstract
Introduction
Adverse drug reactions related to drug–drug interactions cause harm to patients. There is a body of research on signal detection for drug interactions in collections of individual case reports, but limited use in regular pharmacovigilance.
Objective
The aim of this study was to evaluate the feasibility of signal detection of drug–drug interactions in collections of individual case reports of suspected adverse drug reactions.
Methods
This study was conducted in VigiBase, the WHO global database of individual case safety reports. The data lock point was 31 August 2016, which provided 13.6 million reports for analysis after deduplication. Statistical signal detection was performed using a previously developed predictive model for possible drug interactions. The model accounts for an interaction disproportionality measure, expressed suspicion of an interaction by the reporter, potential for interaction through cytochrome P450 activity of drugs, and reported information indicative of unexpected therapeutic response or altered therapeutic effect. Triage filters focused the preliminary signal assessment on combinations relating to serious adverse events with case series of no more than 30 reports from at least two countries, with at least one report during the previous 2 years. Additional filters sought to eliminate already known drug interactions through text mining of standard literature sources. Preliminary signal assessment was performed by a multidisciplinary group of pharmacovigilance professionals from Uppsala Monitoring Centre and collaborating organizations, whereas in-depth signal assessment was performed by experienced pharmacovigilance assessors.
Results
We performed preliminary signal assessment for 407 unique drug pairs. Of these, 157 drug pairs were considered already known to interact, whereas 232 were closed after preliminary assessment for other reasons. Ten drug pairs were subjected to in-depth signal assessment and an additional eight were decided to be kept under review awaiting additional reports. The triage filters had a major impact in focusing our preliminary signal assessment on just 14% of the statistical signals generated by the predictive model for drug interactions. In-depth assessment led to three signals communicated with the broader pharmacovigilance community, six closed signals and one to be kept under review.
Conclusion
This study shows that signals of adverse drug interactions can be detected through broad statistical screening of individual case reports. It further shows that signal assessment related to possible drug interactions requires more detailed information on the temporal relationship between different drugs and the adverse event. Future research may consider whether interaction signal detection should be performed not for individual adverse event terms but for pairs of drugs across a spectrum of adverse events.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Signal detection for drug–drug interactions is more challenging than pairwise drug–adverse drug reaction monitoring, and there has been limited use in regular pharmacovigilance. |
In this study, statistical signal detection combined with triage filters directed signal assessment and ultimately resulted in three communicated signals of drug–drug interactions. |
1 Introduction
Adverse drug reactions (ADRs) related to drug–drug interactions (from here on referred to as drug interactions) constitute an important cause of drug-related problems globally [1,2,3]. Despite modern capability and increased regulatory expectations to investigate potential drug interactions during drug development, interactions between marketed drugs may still be unknown or insufficiently characterized. This not only applies to new drugs entering the market but also to older drugs used in new combinations and to drugs used in new patient populations. Clinical trials are typically not powered to detect drug interactions apart from such that are expected based on prior preclinical and pharmacokinetic studies. Exclusion criteria regarding concurrent medications further reduce their capacity for real-world clinical drug interaction detection. Hence, some drug interactions may only be possible to detect in the postmarketing setting.
Individual case reports of suspected ADRs can be an important source of information of possible adverse reactions resulting from drug interactions [4]. Their coverage is broad, spanning primary and secondary care prescription medicines, as well as over-the-counter and complementary medicines. Reports often relate to individuals who have taken more than one drug at the same time, where some reporters express explicit suspicions that two or more drugs might have interacted to cause the adverse event. Examples of adverse drug interactions detected and analysed in collections of individual case reports include delayed bleeding from oral contraceptives with itraconazole [5], rhabdomyolysis from azithromycin and statins [6], and hyperglycaemia from pravastatin and paroxetine [7].
Early work on methods for statistical interaction surveillance in pharmacovigilance focused on extensions of disproportionality analysis to three-way associations between two drugs and one ADR [5, 8, 9]. The use of association rules has also been proposed [10, 11]. Beyond that, there have been attempts to identify interactions related to specific ADRs based on associated adverse event ‘fingerprints’ [12]. The basis for the study at hand is a previously proposed predictive model that accounts for interaction disproportionality analysis as well as expressed suspicions of an interaction by the reporter, pharmacokinetic plausibility, and reported information indicative of unexpected or altered therapeutic effects [13]. Noguchi et al. provide a recent and comprehensive overview of statistical methods proposed for statistical interaction detection in collections of individual case reports [14]. A broader review of research related to drug–drug interaction signal detection in pharmacovigilance across different data sources is offered by Vilar et al. [15].
Despite available research, evaluations have been retrospective in nature [13] and there is a lack of published initiatives related to signal detection of drug interactions in regular pharmacovigilance. We are not aware of other organizations that use statistical methods to routinely screen collections of individual case reports for suspected adverse drug interactions. This is in stark contrast to the situation for pairwise drug–ADR associations, where disproportionality analysis is in routine use by a wide range of regulators, pharmaceutical companies and academic groups.
The aim of this study was to evaluate the feasibility of signal detection for drug interactions, using a large collection of individual case reports of suspected ADRs. The scope included both pharmacokinetic and pharmacodynamic interactions.
2 Methods
The bulk of this study was carried out in September 2016. Statistical interaction signal detection was combined with triage filters to direct signal assessment. Preliminary signal assessment was performed by a multidisciplinary group of pharmacovigilance professionals over eight dedicated days. Subsequently, in-depth signal assessment was performed by experienced pharmacovigilance assessors.
2.1 VigiBase
The basis for the study were reports of suspected ADRs in VigiBase, the WHO global database of individual case safety reports [16]. Established in 1968, it is the world’s largest and broadest such collection, holding more than 21 million reports from 136 countries (as of November 2019). Reports received into the database until 31 August 2016 were included in the analysis. Suspected duplicate reports were excluded using the vigiMatch algorithm for probabilistic duplicate detection [17]. For technical reasons, deduplication was performed for reports received up until 6 April 2016 only. In total, 13.6 million reports were included in the study.
2.2 Sources of Drug Information
The UK Summary of Product Characteristics (SmPC) in the Electronic Medicines Compendium (eMC) [18], and the US FDA’s drug label in either Drugs@FDA [19] or DailyMed [20] were the primary sources of approved drug information, to establish whether a certain drug–drug adverse event (DDA) interaction could be considered as labelled. The DrugDex [21] and Janusmed interactions [22]Footnote 1 databases were used as complementary sources when relevant information was not found in the first two sources.
2.3 Natural Language Processing of Literature Sources
Automated screening of drug information sources was implemented, and findings on relevant ADRs and interactions were presented to the assessors for each combination. The sources for known ADRs were the PROTECT Database of ADRs listed in the SmPCs of centrally authorised medicinal products within the EU [23], DailyMed, and Martindale. Further sources for known drug interactions were Stockley’s Drug Interaction Alerts [24] and Janusmed. The natural language processing algorithm [25] preprocessed text by stop word removal, stemming and synonym replacement, and then matched to Medical Dictionary for Regulatory Activities (MedDRA®) terms accounting for word permutations and spelling variations. Drug names extracted from the source were mapped to the corresponding drug names in the WHODrug international reference for medicinal product information.
2.4 Statistical Signal Detection for Possible Adverse Drug Interactions
Statistical signal detection for possible adverse drug interactions in VigiBase was performed using a predictive model described by Strandell et al. [13]Footnote 2 and summarized below. In this model, DDA combinations are ranked by probability scores (in the range of 0.04–1.0), where a higher value indicates a higher likelihood that the combinations will be classified as an interaction signal. Table 1 summarizes all predictors and their definitions.
This algorithm includes, as one of its predictors, disproportionality analysis based on the Omega (Ω) statistical interaction measure [8]. Another predictor accounts for whether the two drugs of interest affect the same cytochrome P450 (CYP) enzyme in a way that might lead to an interaction based on the WHODrug Standardised Drug Groupings [26]. Furthermore, it considers expressed suspicions of a drug interaction by the reporter, such as coding the two drugs as ‘interacting’, use of the MedDRA Preferred Term (PT) ‘drug interaction’, and/or inclusion of words starting with ‘interact’ or ‘interakt’ in the case narrative. Reports of altered therapeutic effect are also accounted for, if in combination with (1) specified dosages for both drugs; (2) positive dechallenge intervention (the adverse event abated upon withdrawal of one or both drugs); (3) positive rechallenge intervention (the adverse event reoccurred after one or both drugs were reintroduced); or (4) the two drugs of interest were the only ones listed on the report. Suggestions of unexpected therapeutic response as indicated by the MedDRA PTs ‘therapeutic response unexpected’ and ‘paradoxical drug reaction’ were included as a separate predictor. Finally, there is a predictor accounting for reports of positive dechallenge and overlapping treatment periods when the two drugs of interest were the only ones listed on the report.
We chose this predictive model as the basis for our statistical interaction detection since a retrospective evaluation showed that it performed better in this setting than the Ω interaction disproportionality measure when used on its own [13]. As for the use of Ω, empirical evaluations have shown that statistical interaction models that, like Ω, assume additive effects of non-interacting drugs perform better than those that assume multiplicative effects, such as logistic regression [8, 9].
Disproportionality and CYP are evaluated at the level of the case series. The other predictors are evaluated per report, with scores added together for every report that fulfils a criterion, providing a total contribution score for the case series. With some exceptions specified below, all predictors are computed based only on the reports that listed the two drugs of interest as suspected or interacting, disregarding reports where at least one of the two drugs was listed as concomitant. This includes disproportionality analysis, as well as all but two of the predictors based on the case series. The exceptions are Unexpected therapeutic response and the predictor based on altered therapeutic effect in the presence of solely two drugs included in the report, which count all reports listing the two drugs with the adverse event of interest, including when one or more drug is noted as concomitant, in line with the method proposed in the original publication [13].
The original development and evaluation of the algorithm was based on the WHO Adverse Drug Reaction Terminology (WHO-ART). For the purpose of this study, we mapped WHO-ART PTs used by the algorithm to MedDRA (version 19.0) using the official WHO-ART to MedDRA bridge. In addition, the predictors MedDRA interaction, Unexpected therapeutic response and Altered effect were complemented with relevant PTs containing the words ‘drug’, ‘therapeutic’, ‘level’, ‘increased’, ‘decreased’, ‘clearance’, ‘enzyme’ and ‘treatment’. All included terms are listed in electronic supplementary material 1.
2.5 Triage Filters
Automated filters were applied stepwise with the ambition to restrict the list of DDA combinations to those most likely to be signals for in-depth assessment. An earlier pilot study had indicated that a majority of highlighted DDA combinations reflected already known drug interactions or were due to data quality issues, such as report duplication (unpublished results). To address this, triage filters were targeted at automatically eliminating already known drug interactions and addressing some of the previously identified data quality problems.
The screening was restricted to DDA combinations with no more than 30 reports. This reflected our intent to primarily focus on early signals of drug interactions and to reduce the number of well-known DDA combinations to be assessed.
Beyond that we applied the filters specified in Tables 2 and 3, respectively.
The study design allowed for the implementation of additional (tertiary) filters at one specific time during the course of the 8-day period, with the aim to further focus our assessments. These filters were not prespecified but were designed based on the outcome of the assessments up until that point. Consequently, after about 2 days and having assessed 129 DDA combinations with no signals selected for in-depth assessment, the tertiary filters described in Table 4 were applied in an and/or fashion. In other words, all combinations with either three or more narratives and/or with supporting evidence in Janusmed interactions were selected for preliminary signal assessment.
2.6 Preliminary Signal Assessment
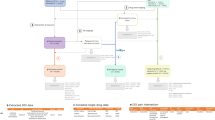
During the preliminary signal assessment step, a group of 21 pharmacovigilance professionals (pharmacists, medical doctors and data scientists) from Uppsala Monitoring Centre and collaborating organizations served as assessors and manually reviewed DDA combinations highlighted by our predictive algorithm and automated triage filters. Their aim was to identify signals that were suitable for in-depth signal assessment. Preliminary signal assessment was performed according to the flowchart shown in Fig. 1.
The first step in the preliminary signal assessment evaluated whether drugs or reported adverse events for each combination were specific enough to support further action. A DDA combination was, for example, deemed non-actionable if the drugs or adverse event were unspecific (e.g. ‘vitamins NOS’ and ‘therapeutic response decreased’), if any of the drugs had been withdrawn from the market, or for other motivations at the assessors’ discretion.
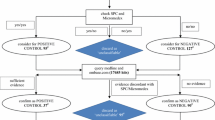
In assessing whether possible drug interactions were already known, a manual review of the information sources specified above was carried out. A drug interaction was considered known when the interaction was mentioned in at least one of the sources of the interacting drugs. In cases where there was information to suggest a drug interaction in DrugDex/Janusmed, but not in the SmPCs/product labels, consultation with one of three designated senior assessors participating in the study was required to judge whether a drug interaction should be considered known. For DDAs where there was no known interaction, the SmPCs/product labels were reviewed to establish whether the adverse event was a known ADR for any or both of the drugs individually. Literature findings and decision-making choices for each manually reviewed combination were documented.
After confirmation by one of the designated senior assessors, DDA combinations whose series of individual case report were assessed as supportive of an interaction were classified as meriting in-depth signal assessment.
Assessors specified the reason for closing signals, e.g. when the DDA combination was non-actionable, the evidence was non-suggestive, or there was a lack of data. An initial analysis of this data suggested heterogeneity in the application of these categories. Results are therefore presented below in aggregate form. Signals that could neither be classified as meriting in-depth assessment or be closed were placed on a monitoring list to keep under review awaiting additional and more informative reports.
2.7 In-Depth Signal Assessment
In-depth assessment was performed by experienced pharmacovigilance assessors, either among the original 21 assessors or within Uppsala Monitoring Centre’s signal review panel.Footnote 3 The details of each in-depth assessment relied on the domain expertise of our individual assessors and the specific circumstances of the signal being assessed; using a common, standardized approach was out of scope for this study. At the end of this process, remaining signals were written, disseminated to the national centres in the WHO Programme for International Drug Monitoring, relevant marketing authorisation holders, and ultimately published in the WHO Pharmaceuticals Newsletter. In case a signal could neither be closed nor support broader dissemination at the time, it was placed on the monitoring list to keep under review awaiting additional reports.
3 Results
3.1 Effect of Triage Filters
The dataset contained 7.4 million DDA combinations, of which 1.6 million contained between 2 and 30 reports. The effect of the triage filters on the top 50,000 DDA combinations, as ranked by the predictive algorithm, is outlined in Fig. 2. The initial filters had approximately the same impact of selecting between 67 and 77% of the top-ranked DDA combinations each. Forty percent of the statistical signals fulfilled all of the initial filters. Table 5 presents the coverage of each secondary filter, and Fig. 3 presents the impact of the tertiary filters.
Overview of the proportions of top-ranked DDA combinations fulfilling our initial filters (40%), remaining after the elimination of DDA combinations by the secondary filters (14%) and being selected by our tertiary filters (6%). All proportions in this graph relate to the 50,000 top DDA combinations as ranked by our predictive model for possible adverse drug interactions. DDA drug–drug adverse event
Venn diagram displaying the proportions of DDA combinations selected by each tertiary filter; in total, 43% of the combinations fulfilled at least one of the criteria (proportions here relate to the number of DDA combinations that remained after the initial filters and secondary filters had been applied). DDA drug–drug adverse event
3.2 Signal Assessment Outcome
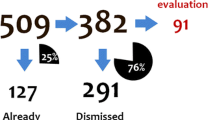
A total of 691 DDA combinations were subjected to preliminary signal assessment, corresponding to 407 unique drug pairs. Since many of the assessed DDA combinations related to the same drug pairs, e.g. 50 related to quetiapine and valproic acid, subsequent results relate to unique drug pairs.
Of the 407 unique drug pairs considered, 157 (39%) were closed as they were related to already known interactions. An additional 232 (57%) were closed for other reasons, such as insufficient or non-suggestive evidence, or that the adverse event was a known ADR for both drugs separately without information to suggest there might be an interaction. Ten drug pairs (2.5%) were classified as signals meriting in-depth assessment, and an additional eight (2.0%) were to be kept under review for additional information. The outcome is presented in Fig. 4.
A list of the 10 signals subjected to in-depth assessment, and their respective outcomes, are available in Table 6. In-depth signal assessments led to three signals for broader dissemination, six closed signals, and one signal to be kept under review. The most common reasons for closing signals after in-depth assessment were a lack of strong individual case reports, lending support to the possible interaction and/or presence of multiple co-reported drugs.
Among the 407 assessed drug pairs, the five most common drugs were acetylsalicylic acid (appearing in 46 drug pairs), valproic acid (24), furosemide (23), fentanyl (14) and metformin (13). The most common PTs were ‘acute kidney injury’ (appearing in 45 drug pairs), ‘drug ineffective’ (42), ‘international normalised ratio increased’ (22), ‘electrocardiogram QT prolonged’ (21) and ‘hyponatraemia’ (19). Preferred Terms related to the MedDRA High Level Group Terms ‘vascular haemorrhagic disorders’, ‘gastrointestinal haemorrhages NEC’, ‘gastrointestinal ulceration and perforation’ and ‘coagulopathies and bleeding diatheses (excl. thrombocytopenic)’ appeared in 76 drug pairs.
3.3 Description of Signals
3.3.1 Ciprofloxacin and Enalapril—Acute Kidney Injury
A case series of 16 reports of acute kidney injury were associated with enalapril and ciprofloxacin, reported as co-suspect or interacting medicines. The suspected interaction was unlabelled but it is known that the use of enalapril and ciprofloxacin separately may lead to renal failure [27, 28]. The signal was supported by a disproportional reporting pattern suggestive of an interaction (Ω025 > 0) and a published epidemiological study. The full signal assessment has been published in the WHO Pharmaceuticals Newsletter [29].
3.3.2 Rosuvastatin and Ticagrelor—Rhabdomyolysis
A case series of five well-documented reports of rhabdomyolysis were associated with ticagrelor and rosuvastatin, reported as co-suspect or interacting medicines. The suspected interaction was unlabelled but it is known that rosuvastatin used on its own may lead to rhabdomyolysis [30]. The signal was supported by a suggestive time-to-onset in three cases where the patients had used rosuvastatin for a long time (month to years) without complaints before ticagrelor was added. The full signal assessment has been published in the WHO Pharmaceuticals Newsletter [31].
3.3.3 Quetiapine and Valproic Acid—Musculoskeletal and Neuropsychiatric Adverse Events
The coadministration of quetiapine and valproic acid is a relatively unexplored option for the acute and maintenance treatment of bipolar disorder. The two drugs are known to interact but the cotreatment is considered well-tolerated [32]. In December 2016, VigiBase held over 1500 reports of quetiapine and valproic acid as co-suspect or interacting medicines. Statistical signals for this pair of drugs were identified for 50 MedDRA PTs. Five of these were selected for in-depth review using vigiPoint: blood creatine phosphokinase increased, coma, depressed level of consciousness, disorientation and rhabdomyolysis [33]. The resulting series of 20 cases was assessed to evaluate the clinical relevance of the interaction. The full signal assessment has been published in the WHO Pharmaceuticals Newsletter [34].
4 Discussion
To our knowledge, this is the first study to evaluate a method for signal detection of drug interactions on a large scale in a regular pharmacovigilance setting. It shows that statistical signal detection for drug interactions is feasible. Ten signals were judged to merit in-depth assessment, and three of these were subsequently published as signals and disseminated to the broader pharmacovigilance community. This was from a pool of 407 evaluated drug pairs, giving a proportion of signals per evaluated drug pair at 0.7%, which is lower than the 1.2% observed historically for pairwise drug–ADR disproportionality analysis in VigiBase, and also substantially so compared with the 3.1% observed with the vigiRank predictive algorithm in the same study [35]. The lower yield may be due to the higher complexity of the causal question in the case of suspected drug interactions. As expected, all signals subjected to in-depth assessment in our study are such that at least one, sometimes both, of the involved drugs are known to cause the adverse event in question on their own. Under such circumstances, there is always a possibility that the observed adverse event is due to one of the drugs and not to a drug interaction, and more detailed information must be available on the reports to assess this possibility. An interaction could, in some cases, be suggested by a time-to-onset pattern where a patient on stable treatment with a drug known to cause the adverse event, experiences the adverse event on the addition of a second drug not known to cause the adverse event [36]. This was the case for the signal on rhabdomyolysis with rosuvastatin and ticagrelor. Similarly, for a dechallenge intervention to be suggestive of a drug interaction, it may need to involve only the drug suspected to have induced the interaction and not the drug known to cause the adverse event itself. If both drugs can cause the adverse event separately, the causal inference is generally difficult and individual cases with strong evidence in favor of a drug interaction are rare.
The basis for our statistical signal detection was a predictive model combining disproportionality analysis with aspects such as possible CYP mediation, suspicion of interactions by the reporter, and more. A previously published retrospective evaluation of this algorithm indicated improved performance compared with disproportionality analysis alone [13], therefore an approach based on just disproportionality analysis may yield fewer signals. It would be relevant to explore further improvements to the algorithm considered in this study, such as to combine the three predictors related to reporters’ suspicions of an interaction. An automated identification of reports with time-to-onset patterns or dechallenge interventions suggestive of drug interactions could be valuable. The predictive model used in vigiRank for pairwise drug–ADR associations [37] is similar in nature but was developed more recently and includes methodological sophistication that could be adopted for improved interaction signal detection. Examples of possible improvements include predictors relating to geographic spread or report quality, and transformations which ensure that after a certain point, additional reports with the same feature do not add to the total score of the combination; for example, if there are already eight reports on an adverse event with the two drugs listed as ‘interacting’, then one more such report would have no impact. This could limit the tendency of the current algorithm to highlight very large report series, which may in turn allow the scope of screening to be expanded to report series of any size. The limit at a maximum of 30 reports used in this study is arbitrary and will lead to missed signals. It has historically been used to reduce the proportion of known ADRs in signal detection and of larger case series that may be difficult to review. However, analyses of associations such as the one for quetiapine and valproic acid presented above can be fruitful, even though they take more time and may require new best practices and computational support to be developed.
The relevant causal question in interaction assessment usually relates to whether two drugs interact in a clinically relevant way, which, if so, could give rise to a range of ADRs. In our study, analyses at the DDA combination level were complicated by single report series resulting in large numbers of statistical signals relating to the same two drugs. An interesting area of future research is whether it may be more effective to perform drug interaction screening directly at the level of drug pairs. Such an approach could account for patterns across different adverse events, possibly looking for stronger associations with ADRs that have a pronounced dose–response relationship. It would present several interesting methodological opportunities and challenges and could potentially yield a substantial increase in performance.
Automated triage filters were an essential component of our approach, focusing our attention on just 14% of the highly ranked DDA combinations. Specifically, more than half were eliminated by one of the filters targeted at already known interactions, and, even so, preliminary signal assessment classified 39% of the drug pairs as already known interactions. One reason for this high proportion of known drug interactions may be that the predictive model used for statistical interaction detection lends substantial weight to reports indicating that the reporter suspected an interaction, and there is evidence to suggest that reporters may be more inclined to do so for known interactions [5]. At the same time, the three published signals from our study were all identified in part because of reports where reporters had conveyed suspicions of drug interactions.
Our study was performed in VigiBase (the world’s largest collection of individual case reports), and while scope and size are important to capture enough reports to enable interaction detection, the amount of information on individual reports is even more critical. Most of the predictors in our algorithm are based on individual reports (e.g. reports of altered or unexpected therapeutic effect, and notes of suspected drug interactions by the reporter) or on the nature of the two drugs (whether there is possible CYP mediation), so they do not rely on broad background data in the way that disproportionality analysis does. In view of this, we believe that the in-house databases of national regulatory authorities and pharmaceutical companies should be fertile ground for interaction signal detection, whereas databases with more limited information on individual reports, such as the public access version of the US FDA’s Adverse Event Reporting System (FAERS) database, may prove less useful for this purpose.
In addition to the limitations highlighted above, it is important to note that this study, by design, focuses on the ability of the proposed approach to detect interaction signals. It shows that such signals can be identified and offers insights into the number of statistical signals that needed to be assessed to detect a signal worthy of broader communication. However, it does not indicate the sensitivity of the proposed approach, i.e. how many drug–drug interactions it may fail to detect. Therefore, further research is required to determine to what extent signal detection in individual case reports may need to be complemented by other approaches to effectively screen for unknown drug interactions in regular pharmacovigilance.
5 Conclusions
This study shows that signals of adverse drug interactions can be detected through broad statistical screening of individual case reports. It further shows that signal assessment related to possible drug interactions requires more detailed information on the temporal relationship between different drugs and the adverse event. Future research may consider whether interaction signal detection should be performed not for individual adverse event terms but for pairs of drugs across a spectrum of adverse events.
Notes
Previously known as the Swedish Finnish Interaction X-referencing (SFINX) database.
Referred to as the lean manual triage in the publication.
References
Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, et al. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ. 2004;329(7456):15–9.
Montane E, Arellano AL, Sanz Y, Roca J, Farre M. Drug-related deaths in hospital inpatients: a retrospective cohort study. Br J Clin Pharmacol. 2018;84(3):542–52.
Becker ML, Kallewaard M, Caspers PW, Visser LE, Leufkens HG, Stricker BH. Hospitalisations and emergency department visits due to drug-drug interactions: a literature review. Pharmacoepidemiol Drug Saf. 2007;16(6):641–51.
van Puijenbroek EP, Egberts ACG, Heerdink ER, Leufkens HGM. Detecting drug–drug interactions using a database for spontaneous adverse drug reactions: an example with diuretics and non-steroidal anti-inflammatory drugs. Eur J Clin Pharmacol. 2000;56(9):733–8.
Van Puijenbroek EP, Egberts ACG, Meyboom RHB, Leufkens HGM. Signalling possible drug–drug interactions in a spontaneous reporting system: delay of withdrawal bleeding during concomitant use of oral contraceptives and itraconazole. Br J Clin Pharmacol. 1999;47(6):689–93.
Strandell J, Bate A, Hagg S, Edwards IR. Rhabdomyolysis a result of azithromycin and statins: an unrecognized interaction. Br J Clin Pharmacol. 2009;68(3):427–34.
Tatonetti N, Denny J, Murphy S, Fernald G, Krishnan G, Castro V, et al. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther. 2011;90(1):133–42.
Norén GN, Sundberg R, Bate A, Edwards IR. A statistical methodology for drug–drug interaction surveillance. Stat Med. 2008;27(16):3057–70.
Thakrar BT, Grundschober SB, Doessegger L. Detecting signals of drug-drug interactions in a spontaneous reports database. Br J Clin Pharmacol. 2007;64(4):489–95.
Harpaz R, Chase HS, Friedman C. Mining multi-item drug adverse effect associations in spontaneous reporting systems. BMC Bioinform. 2010;11(Suppl 9):S7.
Ibrahim H, Saad A, Abdo A, Sharaf EA. Mining association patterns of drug-interactions using post marketing FDA's spontaneous reporting data. J Biomed Inform. 2016;60:294–308.
Lorberbaum T, Sampson KJ, Woosley RL, Kass RS, Tatonetti NP. An integrative data science pipeline to identify novel drug interactions that prolong the QT interval. Drug Saf. 2016;39(5):433–41.
Strandell J, Caster O, Hopstadius J, Edwards IR, Norén GN. The development and evaluation of triage algorithms for early discovery of adverse drug interactions. Drug Saf. 2013;36(5):371–88.
Noguchi Y, Tachi T, Teramachi H. Review of statistical methodologies for detecting drug–drug interactions using spontaneous reporting systems. Front Pharmacol. 2019;10:1319.
Vilar S, Friedman C, Hripcsak G. Detection of drug–drug interactions through data mining studies using clinical sources, scientific literature and social media. Brief Bioinform. 2018;19(5):863–77.
Lindquist M. VigiBase, the WHO global ICSR database system: basic facts. Drug Inform J. 2008;42(5):409–19.
Norén GN, Orre R, Bate A, Edwards IR. Duplicate detection in adverse drug reaction surveillance. Data Min Knowl Disc. 2007;14(3):305–28.
DataPharm. Electronic Medicines Compendium. 2019 [cited 2019]. Available at: https://www.medicines.org.uk/emc
Food and Drug Administration. Drugs@FDA: FDA Approved Drug Products. 2019 [cited 2019]. Available at: https://www.accessdata.fda.gov/scripts/cder/daf/
U.S. National Library of Medicine. DailyMed. 2019 [cited 2019]. Available at: https://dailymed.nlm.nih.gov/dailymed/
IBM® Watson Health™. Micromedex® Pharmaceutical Knowledge, DRUGDEX®. 2019 [cited 2019]. Available at: https://www.micromedexsolutions.com/home/dispatch/ssl/true
Bottiger Y, Laine K, Andersson ML, Korhonen T, Molin B, Ovesjo ML, et al. SFINX-a drug–drug interaction database designed for clinical decision support systems. Eur J Clin Pharmacol. 2009;65(6):627–33.
PROTECT Work Packages 3.1. Reference Dataset of adverse drug reactions up to 12–2011. 2011 [cited 2019]. Available at: https://www.imi-protect.eu/methodsRep.shtml
Baxter K. Stockley’s Drug Interactions, up to 03–2014. 2014 [cited 2019]. Available at: https://www.new.medicinescomplete.com/
Dahlberg G. Implementation and evaluation of a text extraction tool for adverse drug reaction information [thesis]. Uppsala University; 2010.
Lagerlund O, Strese S, Fladvad M, Lindquist M. WHODrug: a global, validated and updated dictionary for medicinal information. Ther Innov Regul Sci. 2020. https://doi.org/10.1007/s43441-020-00130-6.
Electronic Medicines Compendium (EMC). Summary of Product Characteristics (SmPC). Enalapril Maleate Tablets, 10 mg. 2019 [cited 2019]. Available at: https://www.medicines.org.uk/emc/product/2672/smpc
Electronic Medicines Compendium (EMC). Summary of Product Characteristics (SmPC). Ciprofloxacin, 250mg film-coated tablets. 2019 [cited 2019]. Available at: https://www.medicines.org.uk/emc/product/7256/smpc
Savage R. Ciprofloxacin, enalapril and acute kidney injury: strengthening of a drug interaction signal. WHO Pharmaceut Newsl. 2018;1:16–211.
Electronic Medicines Compendium (EMC). Summary of Product Characteristics (SmPC). Crestor, 10mg film-coated tablets. 2019 [cited 2019]. Available at: https://www.medicines.org.uk/emc/product/7559/smpc
Macolić ŠV. Interaction between rosuvastatin and ticagrelor resulting in rhabdomyolysis. WHO Pharmaceut Newsl. 2018;3:10–4.
Electronic Medicines Compendium (EMC). Summary of Product Characteristics (SmPC). SEROQUEL, 300 mg film-coated tablets. 2019 [cited 2019]. Available at: https://www.medicines.org.uk/emc/product/7547/smpc
Juhlin K, Star K, Norén GN. A method for data-driven exploration to pinpoint key features in medical data and facilitate expert review. Pharmacoepidemiol Drug Saf. 2017;26(10):1256–65.
Sartori D. Quetiapine and valproic acid interactions: signal strengthening. WHO Pharmaceut Newsl. 2018;2:19–25.
Caster O, Sandberg L, Bergvall T, Watson S, Norén GN. vigiRank for statistical signal detection in pharmacovigilance: first results from prospective real-world use. Pharmacoepidemiol Drug Saf. 2017;26(8):1006–100.
Strandell J, Norén GN, Hägg S. Key elements in adverse drug interaction safety signals. Drug Saf. 2013;36(1):63–70.
Caster O, Juhlin K, Watson S, Norén GN. Improved statistical signal detection in pharmacovigilance by combining multiple strength-of-evidence aspects in vigiRank. Drug Saf. 2014;37(8):617–28.
Acknowledgements
The authors are indebted to the national centres that make up the WHO Programme for International Drug Monitoring and provide reports to VigiBase. However, the opinions and conclusions of this study are not necessarily those of the various centres or of the WHO. Ola Caster, Pia Caduff-Janosa and Thomas Bradley provided valuable comments on the design and implementation of this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No funding was received to conduct this study.
Conflict of Interest
Sara Hult, Daniele Sartori, Tomas Bergvall, Sara Hedfors Vidlin, Birgitta Grundmark, Johan Ellenius and G. Niklas Norén have no conflicts of interest that are directly relevant to the contents of this study.
Data Sharing
The datasets generated and analysed during the current study are not publicly available due to agreements between contributors of data to the database used (VigiBase) and the custodian of this database. National centres (mainly national drug regulatory authorities) constituting the WHO Programme for International Drug Monitoring contribute data to VigiBase, and Uppsala Monitoring Centre is the custodian in its capacity as the WHO Collaborating Centre for International Drug Monitoring. Some subsets of the data may be available from the corresponding author upon reasonable request.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Hult, S., Sartori, D., Bergvall, T. et al. A Feasibility Study of Drug–Drug Interaction Signal Detection in Regular Pharmacovigilance. Drug Saf 43, 775–785 (2020). https://doi.org/10.1007/s40264-020-00939-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40264-020-00939-y