Abstract
Introduction and Objective
Social media has been suggested as a source for safety information, supplementing existing safety surveillance data sources. This article summarises the activities undertaken, and the associated challenges, to create a benchmark reference dataset that can be used to evaluate the performance of automated methods and systems for adverse event recognition.
Methods
A retrospective analysis of public English-language Twitter posts (Tweets) was performed. We sampled 57,473 Tweets out of 5,645,336 Tweets created between 1 March, 2012 and 1 March, 2015 that mentioned at least one of six medicinal products of interest (insulin glargine, levetiracetam, methylphenidate, sorafenib, terbinafine, zolpidem). Products, adverse events, indications, product-event combinations, and product-indication combinations were extracted and coded by two independent teams of safety reviewers.
Results
The benchmark reference dataset consisted of 1056 positive controls (“adverse event Tweets”) and 56,417 negative controls (“non-adverse event Tweets”). The 1056 adverse event Tweets contained 1396 product-event combinations referring to personal adverse event experiences, comprising 292 different MedDRA® Preferred Terms. The 1171 product-event combinations (83.9%) were confined to four MedDRA® System Organ Classes. The 195 Tweets (18.5%) contained indication information, comprising 25 different Preferred Terms.
Conclusions
A manually curated benchmark reference dataset based on Twitter data has been created and is made available to the research community to evaluate the performance of automated methods and systems for adverse event recognition in unstructured free-text information.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
A manually curated benchmark reference dataset containing positive and negative controls has been created and is made available to the research community |
The benchmark reference dataset can be used to evaluate the performance of automated methods and systems for adverse event recognition in unstructured free-text information |
For the six substances investigated in this study, overall, Twitter posts were mainly about drug ineffectiveness, nervous system/psychiatric disorders, or usage problems (e.g. intentional product misuse). Although the limitation to six substances might limit the generalisability of the dataset, it could provide deeper insights into the real-life usage of these medicinal products and in the use of Twitter regarding adverse events |
1 Introduction
Traditional methods of safety signal detection in licensed pharmaceutical products rely on patients and healthcare professionals to report suspected adverse drug reactions (ADRs) to regulatory agencies or the pharmaceutical companies. Significant under-reporting is well known [1] despite successive efforts to increase reporting. The vast and ever-increasing online presentation of unstructured human experience in social media and a corresponding growth of new technologies offer the opportunity to collect patient perspectives of medication use that might not be otherwise communicated, as well as, at least in theory, the possibility to detect previously unknown ADRs sooner than by traditional methods.
The Innovative Medicines Initiative (IMI) Web-Recognizing Adverse Drug Reactions (WEB-RADR) project was a European Union-funded 3-year project designed to recommend policies, frameworks, tools and methodologies to support reporting of ADRs through mobile applications and the identification of ADRs from social media. Several WEB-RADR activities targeted the acquisition of new insights into drug safety not available with established pharmacovigilance methods. One of the IMI WEB-RADR Work Packages focused on methods to enable signal detection in social media [2]. This work included researching statistical methods for signal detection and methods to enhance automated detection of adverse events (AEs) via entity recognition and mapping of medicinal product and AE terms.
Dozens of automated AE recognition systems have been developed over the past 10 years, with great variability in methodology (e.g. rule based, machine learning, manual curation), source data (e.g. Twitter, Reddit, health-related forums), task solved (e.g. AE span detection, relation extraction, classification of posts as containing AE-related information) and reported performance. We recommend Tricco et al. for a detailed review of those systems [3]. In their article, Tricco et al. state that a direct comparison of the systems is hard to perform, owing to the scarcity of publicly available datasets. However, recent efforts have been made in this direction. In 2017, a shared task on the classification and normalisation of health-related text from social media was performed at the Social Media Mining for Health workshop, involving three subtasks: (1) ADR detection; (2) medication intake classification; and (3) normalisation of ADR expressions [4]. Datasets with training and validation examples were given to teams to train and test their systems, and the final evaluation was made using hold-out hidden test samples, allowing for a fair comparison of performance between the 55 system runs from the 13 participating teams.
Despite its great value for training AE recognition systems, the Social Media Mining for Health dataset cannot be used directly to solve the comprehensive task of finding the products and events mentioned in the text, map them to terminologies, and classify the association between the products and AEs in the given social media text. The Social Media Mining for Health subtask 1 focused on the classification of posts as containing an AE or not, subtask 3 focused on the mapping of short text extracts to MedDRA Preferred Terms (PTs), while the classification of the association between the products and the AEs was not assessed at all. In contrast, the CADEC [5] and the TwiMed [6] corpora are both resources that can be used to train systems to perform the entire AE recognition task described above.
In the ‘ADR Recognition’ work package of IMI WEB-RADR, a large dataset of manually curated Twitter posts (Tweets) was created to aid identification of medicinal product names, AEs, indications, and their associations in social media and to establish a dataset to act as a benchmark reference for method evaluation and comparison. Not intended for training, but rather testing, of AE recognition systems, this dataset represents a novel resource to evaluate the performance of such systems and should be useful to provide measures of their usability when applied to new data.
This article describes the approach taken for selection, collection, sampling, annotation, and quality assurance, and provides descriptive statistics and characteristics of this ‘benchmark reference dataset’. The dataset is publicly available for download [see the Electronic Supplementary Material (ESM) 1].
2 Methods
2.1 Product Selection
The activities described in this article were part of the WEB-RADR project. The WEB-RADR consortium decided to focus these activities on six “drugs of interests” (DOIs), i.e. substances that are manufactured by one of the companies participating in the WEB-RADR consortium (Bayer, Novartis, Sanofi, UCB). For each DOI, a list of product search terms was created using the WHO Drug Global lexicon of global drug names. The product search terms included the products’ generic names, trade names, abbreviations and common misspellings. This resulted in a list of 880 product search terms (between 12 and 418 per DOI) that were used for the Twitter data extraction. Key characteristics of the six DOIs are given in Table 1.
2.2 Twitter Data Extraction, Deduplication and Sampling
The social media data analysed in this report were acquired via an Application Programming Interface from publicly available English-language Twitter posts (Tweets) created between 1 March, 2012 and 1 March, 2015. At the time of data acquisition, Tweets were limited to 140 characters. The data retrieval query that was used to extract data from Twitter contained the 880 product search terms identified in the product selection phase (see Sect. 2.1) and yielded a total of 5,645,336 Tweets. Each of these Tweets potentially contained at least one of the DOIs but not necessarily an AE. The review and annotation of the Tweets later revealed that some Tweets did not contain any DOIs but were included in the data extract as they matched product search terms with alternative connotations (e.g. “ambien”, “concentra”, “freederm”, “intermezzo”).
To remove potentially redundant data, locality-sensitive hashing [7] was applied to the 5,645,336 Tweets resulting in the removal of approximately 80% Tweets identified as duplicates or near-duplicates. The largest single cluster of duplicate Tweets identified by this method contained around 11,000 near-identical Tweets, mostly re-tweets. The remaining subset contained approximately 1.1 million Tweets, and these were grouped by substance name.
From this subset of Tweets, posts were randomly sampled until a target number of at least 1500 posts per DOI was reached. The resulting dataset contained a total of 57,473 Tweets (1–2228 Tweets per product search term). Figure 1 shows the selection and filtering of Tweets through the data extraction, deduplication and sampling process.
2.3 Indicator Score
The Tweets selected in the previous step (see Sect. 2.2) underwent classification by a Bayesian classifier that was previously developed by Epidemico, Inc. (now part of Booz Allen Hamilton) for mining AE discussions in social media data [8], based on Robinson’s method for filtering e-mail spam [9]. The classifier has been trained to identify vernacular language that may describe a suspected ADR or resembles an AE (sometimes referred to as a “Proto-AE”) and calculates an indicator score with values from 0.0 to 1.0. The score indicates the probability that a social media post contains at least one AE (0: low probability, 1: high probability). A penalty of 0.2 is deducted from the indicator score if the post does not contain any identifiable symptom [8].
To avoid any bias on the manual annotation of the Tweets (see Sect. 2.4), the indicator score was not shown to the annotators and was also not used to define the order in which the Tweets went into the annotation process. However, the indicator score was used to define the route a Tweet took through the annotation and quality assurance processes. This is described in Sects. 2.4.4 and 2.4.5.
2.4 Annotation
2.4.1 Setting up the Annotation Environment
To facilitate human review and annotation of the Twitter data, a graphical user interface was developed (Insight Explorer) [10]. Two separate environments were set up, each with a copy of the 57,473 Tweets, to allow two teams to annotate the Tweets independently and in parallel.
2.4.2 Annotation Guideline, Teams and Training
Before the annotation of Tweets started, an annotation guideline was developed that included guidance on how to distinguish between “AE Tweets” and “Non-AE Tweets” and how to extract and code medicinal products and AEs. Two independent teams of annotators were created. Each team (nine people in total per team) worked in one of the annotation environments, and could not see the annotations made by members of the other annotation team. The members of the teams were pharmacovigilance experts with experience in processing individual case safety reports, including coding of medicinal products and AEs.
Each annotator was trained in the annotation guideline and in the use of the tool used to perform the annotation (Insight Explorer). Weekly meetings were held to support the annotators in case of post-training questions regarding the annotation tool, the annotation guideline, or Tweets containing inconclusive or ambiguous content.
2.4.3 Essentials of the Annotation Guideline
Each Tweet was evaluated as an independent Tweet. Therefore, other Tweets from the same user, related Tweets from other users (re-Tweets or replies) or information outside of the Twitter dataset pointing to hyperlinks within the Tweets were not considered for annotation.
Tweets with at least one DOI and at least one AE reported as a personal experience associated with the reported DOI(s) were classified as “AE Tweets”. In those Tweets, all identifiable DOIs and AEs were extracted and mapped to standard dictionary terms, i.e. product name as reported and International Nonproprietary Name for products, and MedDRA PTs for AEs and indications. Furthermore, details about product-event combinations and product-indication combinations, e.g. causal attribution, were evaluated. If a Tweet contained multiple AEs, it was assumed that the AEs occurred over the same period unless the Tweet contained useable information to the contrary.
Tweets containing at least one DOI but no AE, or a DOI with no AE reported as a personal experience, were classified as “Non-AE Tweets”. Tweets without any DOI were also classified as Non-AE Tweets. For Non-AE Tweets, the DOIs, non-DOI products, AEs and indications were not annotated or mapped to standard dictionary terms.
Please note Due to Twitter’s policy, we are not allowed to publish the complete Tweet contents. Therefore, for demonstration purposes in this article, original substance names were substituted by “<substance name>”. In ESM 1 of the online version of this article, the completely evaluated benchmark reference dataset is available, but without the Tweets’ content. Please use the Twitter ID and available programmes (see the link listed in ESM 1) for accessing the Tweets’ content.
Example of an “AE Tweet” and its annotation:
”my doc wanted to give me < substance name 1 > . I said no because I knew I would like it too much. Tried < substance name 2 > but I was sleepwalking/amnesia”
In this example, only < substance name 2 > was identified as a DOI and, therefore, only data for this substance were subsequently annotated. Of note, even if < substance name 1 > would have been a DOI, the Tweet does not contain a personal experience of an AE associated with < substance name 1 > and hence, no product-event combination would have been annotated for it.
Annotation result:
Classification: AE Tweet
Product(s) as reported: < substance name 2>
Product coded (International Nonproprietary Name): < substance name 2 coded>
Event(s) as reported: amnesia; sleepwalking
Event(s) coded (PT): Amnesia; Somnambulism
Product event(s): < substance name 2 > : Amnesia; < substance name 2 > : Somnambulism
Indication(s) as reported:
Indication(s) coded (PT):
Product indication(s):
Please note: In this example, no indication is reported. Therefore, those fields are left blank.
Two typical examples of “Non-AE Tweets”:
”? < substance name > is a pill that works through the bloodstream to target and attack the infection at its source underneath the nail.?“
“<substance name > , which was priced at Rs 2.28 lakh per month is now available for Rs 6,600“
2.4.4 Annotation Process
The annotation process is outlined in Fig. 2. The two different Insight Explorer database instances are labelled as “IE#1” and “IE#2”. The indicator scores of the Tweets were not displayed to the annotation teams to avoid bias on their manual annotation.
The original goal was that all 57,473 Tweets would be reviewed manually by the two annotation teams. However, it was found that the annotation took longer than anticipated and would not be completed within the timeline defined by the WEB-RADR project. Hence, an exploratory analysis was performed to investigate the potential for annotation automation of “Non-AE Tweets”.
At the time of the exploratory analysis, 15,195 Tweets had been reviewed and, within those, 91 “AE Tweets” had been identified. For Tweets with an indicator score below 0.3, only five AE Tweets were found compared with 5982 Non-AE Tweets. Based on this finding, it was determined that Tweets with an indicator score below 0.3 could be considered Non-AE Tweets and be excluded from manual annotation without significant loss of precision and recall. Applying this filter to the entire dataset of 57,473 Tweets resulted in the classification of 24,311 Tweets as Non-AE Tweets, leaving 33,162 Tweets for manual human curation.
The 33,162 Tweets with an indicator score ≥ 0.3 were manually curated by the two independent annotation teams. Both annotation teams agreed on the classification of 31,340 Non-AE Tweets and 507 AE Tweets (see Fig. 2). For 1315 Tweets, the classification by the two annotation teams differed, illustrating the difficulty of interpreting the content of Tweets (see Sect. 4 for details).
Tweets with indicator scores between 0.3 and 0.7 and classified by both teams as Non-AE Tweets were not processed any further (n = 30,303). For the remaining Tweets with an indicator score ≥ 0.3 (n = 2859), a 100% quality control was performed by a team of experienced MedDRA coders to propose the annotations for the benchmark reference dataset. As shown in Fig. 2, these 2859 Tweets comprised the concordantly classified Non-AE Tweets with an indicator score ≥ 0.7 (n = 1037), the discordantly classified Tweets with an indicator score ≥ 0.3 (n = 1315) and the concordantly classified AE Tweets (n = 507). This quality control process resulted in the identification of 991 AE Tweets. Finally, two quality assurance measures were performed to make final refinements to the benchmark reference dataset.
2.4.5 Quality Assurance
Two quality assurance measures were defined and performed to yield the best quality of the benchmark reference dataset under the given circumstances of this project.
Quality Assurance #1 Of 600 randomly selected Tweets (300 AE Tweets and 300 Non-AE Tweets) with an indicator score ≥ 0.3, were independently evaluated by a team not involved in the prior annotation process (see Quality Assurance #1 in Fig. 2). Among the 300 AE Tweets, a total of 46 Tweets with issues were found: non-DOI products were wrongly identified as DOIs (n = 14); AEs were coded to the wrong PT (n = 25); one Tweet was wrongly identified as an AE Tweet (n = 1); and misspellings were identified (n = 6). Among the 300 Non-AE Tweets, eight were found with AEs (i.e. AE Tweets) [2.7%].
The identified issues were resolved in the benchmark reference dataset.
Quality Assurance #2 Tweets were sorted by descending indicator scores, a total of 1200 Non-AE Tweets were assigned to batches of 100 each (n = 12 batches), and a 100% Tweet content check was performed to identify potentially missed additional AE Tweets (see Quality Assurance #2 in Fig. 2). Among all Tweets (both AE Tweets and Non-AE Tweets together) within the range of an indicator score defined by each batch, the proportion of missed AE Tweets (annotated as Non-AE Tweets but identified as AE Tweets in the second quality assurance) was computed. This proportion was found to vary between 0.9% [batch 12: one missed AE Tweet/(100 Non-AE Tweets + 15 AE Tweets)] and 6.7% [batch 2: 12 missed AE Tweets/(100 Non-AE Tweets + 80 AE Tweets)]. In this quality assurance step, a total of 58 additional AE Tweets were identified and annotated, and the benchmark reference dataset updated accordingly.
3 Results
3.1 Characteristics of the Benchmark Reference Dataset
3.1.1 Format and Accessibility
The benchmark reference dataset is publicly available for download in XLSX format in the online version of this article as ESM 1. The file includes a table that describes the content of the dataset, i.e. column names and descriptions.
3.1.2 Positive and Negative Controls
The benchmark reference dataset contains 57,473 Tweets, with 1056 AE Tweets (1.8%; positive controls) and 56,417 Non-AE Tweets (98.2%; negative controls).
3.1.3 Effect of the Quality Control and Quality Assurance Measures
The comparison of the benchmark reference dataset with the dataset from before the quality steps revealed the following: from the discordantly classified Tweets (n = 1315), and the concordantly classified AE Tweets (n = 507), the quality control process resulted in the identification of 991 AE Tweets. In quality assurance step #1, one Non-AE Tweet was removed and eight AE Tweets were added. In step #2, 58 additional AE Tweets were added, resulting in total of 1056 AE Tweets.
3.1.4 Number of Adverse Events (AEs) per AE Tweet
The AE Tweets in the benchmark reference dataset contain between one and eight AEs per Tweet: 74.6% contain one AE, 20.1% two AEs, 4.3% three AEs, 0.9% four AEs, 0.1% five AEs and 0.1% eight AEs. Thus, around 95% of all AE Tweets contain a maximum of two AEs. This distribution of AEs per Tweet is comparable to results published by Patel et al. [11].
3.2 Distribution of Substances in Tweets
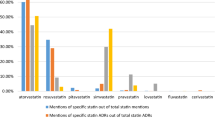
Within the 57,473 Tweets of benchmark reference dataset, 80,429 product search terms were found (see Table 1). There were 7704 Tweets with more than one product search term and 403 Tweets with two different DOIs. About 76% of all Tweets contained the two most frequently mentioned DOIs: terbinafine (24,781 Tweets, 42.8%) and zolpidem (19,403 Tweets, 33.5%).
The analysis of the 1056 AE Tweets shows that the DOIs are heterogeneously distributed (see Table 1). At the lower end, sorafenib was only mentioned in 14 AE Tweets (1.3%), whereas at the upper end, methylphenidate was mentioned in 357 Tweets (33.8%). Of the AE Tweets (n = 932), 88.3% refer to the three most mentioned DOIs (methylphenidate, zolpidem, levetiracetam).
An interesting finding is the divergent occurrences of DOIs in the 57,473 Tweets vs the 1056 AE Tweets, most pronounced for methylphenidate and terbinafine: while the percentage increased for methylphenidate from 6.9 to 33.8%, it decreased for terbinafine from 42.8 to 3.5%. The cause of this difference in prevalence in all Tweets vs AE Tweets has not yet been analysed.
3.3 Distribution of Event Terms in AE Tweets
In the 1056 AE Tweets in the benchmark reference dataset, 1396 AEs were identified, annotated and coded to MedDRA PTs comprising 292 different PTs (see ESM 1). A total of 83.9% (n = 1171) of these AEs map to just four primary System Organ Classes (SOCs): SOC General disorders and administration site conditions (37.2%, n = 519), SOC Psychiatric disorders (26.5%, n = 370), SOC Nervous system disorders (11.5%, n = 161), and SOC Injury, poisoning and procedural complications (8.7%, n = 121) [see Table 2; see ESM 2 for the table showing all SOCs].
Table 2 also shows the distributions of the six DOIs in the four most frequently reported SOCs. Except for SOC Nervous system disorder where levetiracetam appears on top of the table, zolpidem and methylphenidate are the most frequent DOIs in the top SOCs.
Table 3 shows the most frequently reported PTs within the top four SOCs. In SOC General disorders and administration site conditions, PT Drug ineffective and rather unspecific AE terms dominate the list. In SOC Psychiatric disorders and SOC Nervous system disorders, PTs hint at the use of psychotropic substances or psychotropic events, respectively. The PTs in SOC Injury, poisoning and procedural complications mainly refer to administration and dose errors and intentional product misuse.
3.4 Distribution of Indications in AE Tweets
Within the 1056 AE Tweets, 195 Tweets (18.5%) contain indication information. One Tweet contains two different indications. Hence, a total of 196 indications were identified in the AE Tweets and coded to MedDRA PTs. In 117 of 195 Tweets (60.0%), indications were identified by Twitter hashtags (e.g. #ADHD) or references (e.g. @epilepsyaction), which comprise 49 different hash tags/references (see ESM 3). In the remaining 78 Tweets (40.0%), indications were identified by the explicit description of the purpose of use, e.g. “I’ve had athletes foot for two years now lol… < substance name > … dont work.”.
Table 4 shows the MedDRA PTs of the 25 different indications that were identified and coded. Please note that one indication (PT Sleep disorder) occurs twice, i.e. for zolpidem and for levetiracetam. The top five indications account for 161 of 196 indications (82.1%), whereas 168 of 196 indications (85.7%) were reported for the top three substances (levetiracetam n = 96, zolpidem n = 37, methylphenidate n = 35).
Of note, the values in column “Potential off-label use” in Table 4 are the result of the comparison of the reported indication against the respective DOI’s Summary of Product Characteristics, not against country-specific labels. An example of a potential off-label use reported in a Tweet is as follows: “We asked < reference > what she does to help her sleep. Her answer? 1,500 mg of < substance name > and a small amount of (legal) cannabis”.
4 Discussion
The process employed to create the benchmark reference dataset, summarised as follows, was designed to achieve the best-possible quality given the time and resources available in this project:
Set up of two independent annotation teams and provision of annotation guidelines
Execution of training for the annotation teams, weekly team meetings and independent issue discussions with both teams
Performing independent quality control and assurance steps
Final review and, as required, revision of the AE Tweets in the benchmark reference dataset by a senior medical case evaluator
However, despite careful planning, we faced organisational challenges because of staff turnover and especially, challenges related to the often-ambiguous content of Tweets that sometimes resulted in discordant interpretation and annotation of Tweets:
The duration of the Tweet annotation was around 10 months (November 2015–September 2016) and the staff turnover within the annotation teams was high. This meant continued efforts in onboarding and training new team members, and a mix of annotators that already had gained experience with Tweet annotation or were new to this task
In social media posts, vernacular language is used, which can be interpreted differently by different people reading the posts
Discrepant identification of DOI due to:
Use of abbreviations (e.g. “zolpi” instead of “zolpidem”)
Use of ambiguous terms (e.g. “intermezzo”)
Use of the same trade names for different medications [e.g. “freederm” (Terbinafine) vs “freederm” (nicotinamide)]
Room for interpretation of the reported AEs as personal experience vs a general statement, e.g.:
“This medication < substance name > every hours makes you hungry”
“<substance name > dont make me hungry leh. but it might be relaxing u from anxiety and thus ur appetite return”
“Ok, Must be immune to < substance name 1 > . < substance name 2 > it is then or < substance name 3 > to you Americans”
The decision for this study to annotate a single Tweet as is (i.e. without reviewing prior or later Tweets of the same Twitter user and without the means to follow-up with the user to clarify what he/she meant by the Tweet) made the annotation susceptible to a high degree of discordant interpretation by the two annotation teams. These discordances were then reviewed and resolved in the quality control and quality assurance steps, as described in Sects. 2.4.4 and 2.4.5.
In terms of product scope, the benchmark reference dataset is limited, as it includes only six different DOIs. Thus, this dataset, used alone, would not be very useful as input for training of AEs and/or indication classification and a mapping system. However, it can be very useful for testing such systems and comparing the performance of different systems. The benchmark reference dataset has, to our knowledge, a unique combination of features that make it a worthwhile addition to existing reference datasets:
It contains both positive and negative controls (AE Tweets and Non-AE Tweets, respectively), which allows calculating performance indicators for a tested system, such as precision and recall
Besides the classification of AE Tweet vs Non-AE Tweet, it also contains the details of all product-event combinations and product-indication combinations identified in each AE Tweet, including verbatims and dictionary mappings for DOIs, AEs and indications. This allows testing of narrow-scoped classifiers and wide-ranged entity recognition and mapping systems
The annotated AEs refer to personal experiences of drug effects with an explicitly reported, or reasonably assumed, timely or causal association between drug use and AE
Both AEs and indications have been annotated and coded to MedDRA. This allows testing of algorithms that are designed to identify indications, or to distinguish between AEs and indications, respectively
On the topic of identifying indications and non-ADR medical conditions in social media, only a small number of publications were found [8, 12,13,14,15] of which one refers to French language [12] and one to Spanish language social media content [15]. Sarker et al. proposed a concept of identifying indications by the frequency of the occurrence of drug-ADR pairs mentioned in close proximity within posts [14], while Nikfarjam et al. [13] reported that the majority of false-positive errors “… were caused by mentions that were confused with indications or non-ADR clinical mentions …”, which indicates that AEs and indications could currently not satisfactorily be separated from each other by automated means and are still a challenge for automated systems and a field for future studies. As the benchmark reference dataset contains both AE and indication annotations and mappings, it should be helpful as a reference for such studies and for improving methods that are capable of identifying both AEs and indications.
When 15,195 Tweets had been reviewed by both annotation teams, we conducted an exploratory analysis and discovered we can use Epidemico’s indicator score < 0.3 to automatically flag Tweets as Non-AE Tweets, accepting the loss of a limited number of potential AE Tweets. Five AE Tweets, which were identified in the exploratory analysis and quality control and assurance steps, had an indicator score even below 0.3; these Tweets mainly contained rare PTs (e.g. PT Maternal exposure during pregnancy, PT Condition aggravated, PT Petit mal epilepsy, PT Cardiotoxicity).
For the detection of AE Tweets, some publications reported an indicator score threshold of ≥ 0.65 or ≥ 0.7 to automatically distinguish between so-called “Proto-AE” (which resembles what is called “AE Tweet” in this article) and Non-AE Tweets [8, 11, 16]. According to our results in the benchmark reference dataset, only 775 of 1056 AE Tweets (73.4%) had an indicator ≥ 0.65, and 662 of 1056 AE Tweets (62.7%) had an indicator score ≥ 0.7. Hence, for the dataset of 57,473 Tweets: applying an indicator score threshold ≥ 0.65 would have missed 26.6% and for a threshold ≥ 0.7, 37.3% of the benchmark AE Tweets. A detailed analysis of the performance of Epidemico’s indicator score against the benchmark reference dataset is beyond the scope of this article and hence, is not included here.
5 Conclusions
The proper identification, extraction and mapping of product-event and product-indication combinations in free text is still a challenge. The IMI WEB-RADR project established a publicly available benchmark reference dataset that can be used to test and compare the performance of entity recognition methods targeted at the automated identification and mapping of personal experiences of AEs and indications reported in social media, especially Twitter. Therefore, it hopefully contributes to the improvement of existing and the development of new methods and systems thus contributing to the advancement of pharmacovigilance.
References
Hazell L, Shakir SA. Under-reporting of adverse drug reactions : a systematic review. Drug Saf. 2006;29(5):385–96.
Caster O, Dietrich J, Kurzinger ML, Lerch M, Maskell S, Noren GN, et al. Assessment of the utility of social media for broad-ranging statistical signal detection in pharmacovigilance: results from the WEB-RADR Project. Drug Saf. 2018;41(12):1355–69.
Tricco AC, Zarin W, Lillie E, Jeblee S, Warren R, Khan PA, et al. Utility of social media and crowd-intelligence data for pharmacovigilance: a scoping review. BMC Med Inform Decis Mak. 2018;18(1):38.
Sarker A, Belousov M, Friedrichs J, Hakala K, Kiritchenko S, Mehryary F, et al. Data and systems for medication-related text classification and concept normalization from Twitter: insights from the Social Media Mining for Health (SMM4H)-2017 shared task. J Am Med Inform Assoc. 2018;25(10):1274–83.
Karimi S, Metke-Jimenez A, Kemp M, Wang C. Cadec: a corpus of adverse drug event annotations. J Biomed Inform. 2015;55:73–81.
Alvaro N, Miyao Y, Collier N. TwiMed: Twitter and PubMed comparable corpus of drugs, diseases, symptoms, and their relations. JMIR Public Health Surveill. 2017;3(2):e24.
Gionis A, Indyk P, Motwani R. Similarity search in high dimensions via hashing. Proceedings of the 25th International Conference on Very Large Data Bases. Morgan Kaufmann Publishers Inc.; 1999: p. 518–29.
Pierce CE, Bouri K, Pamer C, Proestel S, Rodriguez HW, Van Le H, et al. Evaluation of Facebook and Twitter monitoring to detect safety signals for medical products: an analysis of recent FDA safety alerts. Drug Saf. 2017;40(4):317–31.
Robinson G. A statistical approach to the spam problem. Linux J. 2003;2003(107):3.
Casperson TA, Painter JL, Dietrich J. Strategies for distributed curation of social media data for safety and pharmacovigilance. Proceedings of the International Conference on Data Mining (DMIN), The Steering Committee of The World Congress in Computer Science, Computer Engineering and Applied Computing (WorldComp). 2016:118.
Patel RBM, Jani M, Dasgupta N, Winakor C, Nenadic G, Dixon W. Frequent discussion of insomnia and weight gain with glucorticoid therapy: an analysis of Twitter posts. npj Digital Medicine. 2018.
Morlane-Hondère F, Grouin C, Zweigenbaum P. Identification of drug-related medical conditions in social media. Portorož: European Language Resources Association (ELRA); 2016 May: p. 2022–8.
Nikfarjam A, Sarker A, O’Connor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Inform Assoc. 2015;22(3):671–81.
Sarker A, Ginn R, Nikfarjam A, O’Connor K, Smith K, Jayaraman S, et al. Utilizing social media data for pharmacovigilance: a review. J Biomed Inform. 2015;54:202–12.
Segura-Bedmar I, de la Peña González S, Martínez P. Extracting drug indications and adverse drug reactions from Spanish health social media. Baltimore (MD): Association for Computational Linguistics; 2014. p. 98–106.
Powell GE, Seifert HA, Reblin T, Burstein PJ, Blowers J, Menius JA, et al. Social media listening for routine post-marketing safety surveillance. Drug Saf. 2016;39(5):443–54.
Acknowledgements
The authors are indebted to the following colleagues, past or present, within the WEB-RADR consortium who provided technical support that enabled the research presented in this paper: Tim Casperson, Johan Ellenius, Geoffry Gipson, Daniela Victoria Grohmann, Johnny Gunn, Karin Hace, Simon Maskell, Paul Murphy, Victoria Newbould, the Novartis annotation team, Sheila O’Brien, Jeffery Painter, Carrie Pierce, Sue Rees, Erik Scalfaro, Tata Consultancy Services on behalf of the Bayer AG and Miguel Texeira. The opinions and conclusions of this study are not necessarily those of the national centres which make up the WHO Programme for International Drug Monitoring nor of the WHO. MedDRA® (the Medical Dictionary for Regulatory Activities) terminology is the international medical terminology developed under the auspices of the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH). The MedDRA® trademark is owned by the International Federation of Pharmaceutical Manufacturers and Associations on behalf of ICH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
All research presented here was conducted by the authors listed. This collaborative effort is provided via the WEB-RADR project (http://www.web-radr.eu), which is supported by the Innovative Medicines Initiative Joint Undertaking under Grant Agreement No. 115632, resources of which comprise financial contributions from the European Union’s Seventh Framework Programme (FP7/2007-2013) and European Federation of Pharmaceutical Industries and Associations companies’ in-kind contribution (http://www.imi.europa.eu). In addition, general support for the development of the social media listening platform was provided by GlaxoSmithKline, independent of the research presented herein.
Conflict of Interest
Juergen Dietrich, Lucie M. Gattepaille, Britta Anne Grum, Letitia Jiri and Daniele Sartori have no conflicts of interest that are directly relevant to the content of this article. Antoni Wisniewski is an employee of AstraZeneca and shareholder of AstraZeneca and GlaxoSmithKline. Magnus Lerch provided scientific advice and support to Bayer AG within the WEB-RADR project and has received compensation for his work from Bayer AG.
Ethics Approval
All human subject data used in this analysis were publicly available and used in a de-identified format whenever possible.
Data Sharing
Data generated or analysed during this study are included in this published article (and its supplementary information files).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Dietrich, J., Gattepaille, L.M., Grum, B.A. et al. Adverse Events in Twitter-Development of a Benchmark Reference Dataset: Results from IMI WEB-RADR. Drug Saf 43, 467–478 (2020). https://doi.org/10.1007/s40264-020-00912-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40264-020-00912-9