Abstract
Introduction
Given that adverse drug effects (ADEs) have led to post-market patient harm and subsequent drug withdrawal, failure of candidate agents in the drug development process, and other negative outcomes, it is essential to attempt to forecast ADEs and other relevant drug–target–effect relationships as early as possible. Current pharmacologic data sources, providing multiple complementary perspectives on the drug–target–effect paradigm, can be integrated to facilitate the inference of relationships between these entities.
Objective
This study aims to identify both existing and unknown relationships between chemicals (C), protein targets (T), and ADEs (E) based on evidence in the literature.
Materials and Methods
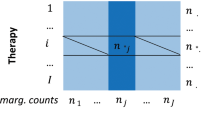
Cheminformatics and data mining approaches were employed to integrate and analyze publicly available clinical pharmacology data and literature assertions interrelating drugs, targets, and ADEs. Based on these assertions, a C–T–E relationship knowledge base was developed. Known pairwise relationships between chemicals, targets, and ADEs were collected from several pharmacological and biomedical data sources. These relationships were curated and integrated according to Swanson’s paradigm to form C–T–E triangles. Missing C–E edges were then inferred as C–E relationships.
Results
Unreported associations between drugs, targets, and ADEs were inferred, and inferences were prioritized as testable hypotheses. Several C–E inferences, including testosterone → myocardial infarction, were identified using inferences based on the literature sources published prior to confirmatory case reports. Timestamping approaches confirmed the predictive ability of this inference strategy on a larger scale.
Conclusions
The presented workflow, based on free-access databases and an association-based inference scheme, provided novel C–E relationships that have been validated post hoc in case reports. With refinement of prioritization schemes for the generated C–E inferences, this workflow may provide an effective computational method for the early detection of potential drug candidate ADEs that can be followed by targeted experimental investigations.






Similar content being viewed by others
Notes
MedDRA® the Medical Dictionary for Regulatory Activities terminology is the international medical terminology developed under the auspices of the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH).
References
Aronson JK. Distinguishing hazards and harms, adverse drug effects and adverse drug reactions: implications for drug development, clinical trials, pharmacovigilance, biomarkers, and monitoring. Drug Saf. 2013;36:147–53.
Shepherd G, Mohorn P, Yacoub K, May DW. Adverse drug reaction deaths reported in United States vital statistics, 1999–2006. Ann Pharmacother. 2012;46:169–75.
Suh DC, Woodall BS, Shin SK, Hermes-De Santis ER. Clinical and economic impact of adverse drug reactions in hospitalized patients. Ann Pharmacother. 2000;34:1373–9.
Agency for Healthcare Research and Quality. Reducing and preventing adverse drug events to decrease hospital costs. AHRQ Archive. https://archive.ahrq.gov/research/findings/factsheets/errors-safety/aderia/ade.html. Accessed 2 Apr 2014.
Ninan B, Wertheimer A. Withdrawing drugs in the U.S. versus other countries. Innov Pharm. 2012;3(3):Article 87. https://pubs.lib.umn.edu/index.php/innovations/article/view/269/263. Accessed 19 Apr 2018.
Swanson DR. Migraine and magnesium: eleven neglected connections. Perspect Biol Med. 1988;31:526–57.
Ramadan NM, Halvorson H, Vande-Linde A, Levine SR, Helpern JA, Welch KM. Low brain magnesium in migraine. Headache. 1989;29:416–9.
US Food and Drug Administration. openFDA datasets: FAERS. https://open.fda.gov/data/faers/. Accessed 19 Apr 2018.
Harpaz R, Haerian K, Chase HS, Friedman C. Statistical mining of potential drug interaction adverse effects in FDA’s spontaneous reporting system. AMIA Annu Symp Proc AMIA Symp. 2010;2010:281–5.
Schuemie MJ, Coloma PM, Straatman H, Herings RMC, Trifirò G, Matthews JN, et al. Using electronic health care records for drug safety signal detection: a comparative evaluation of statistical methods. Med Care. 2012;50:890–7.
Bai JPF, Abernethy DR. Systems pharmacology to predict drug toxicity: integration across levels of biological organization. Annu Rev Pharmacol Toxicol. 2013;53:451–73.
Yang L, Agarwal P. Systematic drug repositioning based on clinical side-effects. PLoS ONE. 2011;6:e28025.
Yamanishi Y, Pauwels E, Kotera M. Drug side-effect prediction based on the integration of chemical and biological spaces. J Chem Inf Model. 2012;52:3284–92.
Cheng F, Li W, Wang X, Zhou Y, Wu Z, Shen J, et al. Adverse drug events: database construction and in silico prediction. J Chem Inf Model. 2013;53:744–52.
Cami A, Arnold A, Manzi S, Reis B. Predicting adverse drug events using pharmacological network models. Sci Transl Med. 2011;3:114ra127.
Chen B, Ding Y, Wild DJ. Assessing drug target association using semantic linked data. PLoS Comput Biol. 2012;8:e1002574.
Campillos M, Kuhn M, Gavin A-C, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–6.
Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–7.
Oprea TI, Nielsen SK, Ursu O, Yang JJ, Taboureau O, Mathias SL, et al. Associating drugs, targets and clinical outcomes into an integrated network affords a new platform for computer-aided drug repurposing. Mol Inform. 2011;30:100–11.
Davis AP, Wiegers TC, Roberts PM, King BL, Lay JM, Lennon-Hopkins K, et al. A CTD-Pfizer collaboration: manual curation of 88,000 scientific articles text mined for drug-disease and drug-phenotype interactions. Database. 2013;2013:bat080.
King BL, Davis AP, Rosenstein MC, Wiegers TC, Mattingly CJ. Ranking transitive chemical-disease inferences using local network topology in the comparative toxicogenomics database. PLoS One. 2012;7:e46524.
Simon Z, Peragovics A, Vigh-Smeller M, Csukly G, Tombor L, Yang Z, et al. Drug effect prediction by polypharmacology-based interaction profiling. J Chem Inf Model. 2012;52:134–45.
Wang Y, Chen S, Deng N, Wang Y. Drug repositioning by kernel-based integration of molecular structure, molecular activity, and phenotype data. PLoS One. 2013;8:e78518.
Wallach I, Jaitly N, Lilien R. A structure-based approach for mapping adverse drug reactions to the perturbation of underlying biological pathways. PLoS One. 2010;5:e12063.
Mathur S, Dinakarpandian D. Drug repositioning using disease associated biological processes and network analysis of drug targets. AMIA Annu Symp Proc. 2011;2011:305–11.
Kim Kjærulff S, Wich L, Kringelum J, Jacobsen UP, Kouskoumvekaki I, Audouze K, et al. ChemProt-2.0: visual navigation in a disease chemical biology database. Nucleic Acids Res. 2013;41:D464–9.
Jacob L, Vert J-P. Protein-ligand interaction prediction: an improved chemogenomics approach. Bioinforma Oxf Engl. 2008;24:2149–56.
Lo HZ, Ding W, Nazeri Z. Mining adverse drug reactions from electronic health records. In: 2013 IEEE 13th Int Conf Data Min Workshop. 2013. p. 1137–40.
Jensen K, Soguero-Ruiz C, Mikalsen KO, Lindsetmo R-O, Kouskoumvekaki I, Girolami M, et al. Analysis of free text in electronic health records for identification of cancer patient trajectories. Sci Rep. 2017;7:46226.
Yadav P, Steinbach M, Kumar V, Simon G. Mining electronic health records (EHRs): a survey. ACM Comput Surv. 2018;50:85:1–85:40.
Huang R, Southall N, Wang Y, Yasgar A, Shinn P, Jadhav A, et al. The NCGC Pharmaceutical Collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci Transl Med. 2011;3:80ps16. https://tripod.nih.gov/npc/. Accessed 23 May 2018.
Fourches D, Muratov E, Tropsha A. Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J Chem Inf Model. 2010;50:1189–204.
Fourches D, Muratov E, Tropsha A. Curation of chemogenomics data. Nat Chem Biol. 2015;11:535.
Fourches D, Muratov E, Tropsha A. Trust, but verify II: a practical guide to chemogenomics data curation. J Chem Inf Model. 2016;56:1243–52.
Kuz’min VE, Artemenko AG, Muratov EN. Hierarchical QSAR technology based on the Simplex representation of molecular structure. J Comput Aided Mol Des. 2008;22:403–21.
Yang H, Qin C, Li YH, Tao L, Zhou J, Yu CY, et al. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016;44:D1069–74.
UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–12.
Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6:343.
National Library of Medicine. Unified Medical Language System (UMLS). 2013. https://www.nlm.nih.gov/research/umls/. Accessed 2 Mar 2015.
Kuhn M, Szklarczyk D, Franceschini A, von Mering C, Jensen LJ, Bork P. STITCH 3: zooming in on protein–chemical interactions. Nucleic Acids Res. 2012;40:D876–80.
Davis AP, Murphy CG, Johnson R, Lay JM, Lennon-Hopkins K, Saraceni-Richards C, et al. The comparative toxicogenomics database: update 2013. Nucleic Acids Res. 2013;41:D1104–14.
Baker NC, Hemminger BM. Mining connections between chemicals, proteins, and diseases extracted from Medline annotations. J Biomed Inform. 2010;43:510–9.
MedDRA. 2013. https://www.meddra.org/. Accessed 19 Apr 2018.
Wohlgemuth G, Haldiya PK, Willighagen E, Kind T, Fiehn O. The Chemical Translation Service—a web-based tool to improve standardization of metabolomic reports. Bioinformatics. 2010;26:2647–8.
Cover TM, Thomas JA. Relative entropy and mutual information. In: Elements of information theory. 2nd ed. New York: Wiley Interscience; 2006. p. 19–20.
Wysowski DK, Swartz L. Adverse drug event surveillance and drug withdrawals in the United States, 1969-2002: the importance of reporting suspected reactions. Arch Intern Med. 2005;165:1363–9.
Trifirò G, Pariente A, Coloma PM, Kors JA, Polimeni G, Miremont-Salamé G, et al. Data mining on electronic health record databases for signal detection in pharmacovigilance: which events to monitor? Pharmacoepidemiol Drug Saf. 2009;18:1176–84.
Houwerzijl EJ, Blom NR, van der Want JJL, Esselink MT, Koornstra JJ, Smit JW, et al. Ultrastructural study shows morphologic features of apoptosis and para-apoptosis in megakaryocytes from patients with idiopathic thrombocytopenic purpura. Blood. 2004;103:500–6.
Mitra D, Kim J, MacLow C, Karsan A, Laurence J. Role of caspases 1 and 3 and Bcl-2-related molecules in endothelial cell apoptosis associated with thrombotic microangiopathies. Am J Hematol. 1998;59:279–87.
Taler M, Gil-Ad I, Lomnitski L, Korov I, Baharav E, Bar M, et al. Immunomodulatory effect of selective serotonin reuptake inhibitors (SSRIs) on human T lymphocyte function and gene expression. Eur Neuropsychopharmacol. 2007;17:774–80.
Metjian A, Abrams CS. New advances in the treatment of adult chronic immune thrombocytopenic purpura: role of thrombopoietin receptor-stimulating agents. Biol Targets Ther. 2009;3:499–513.
Schipperus M, Fijnheer R. New therapeutic options for immune thrombocytopenia. Neth J Med. 2011;69:480–5.
Metcalfe PD, Leslie JA, Campbell MT, Meldrum DR, Hile KL, Meldrum KK. Testosterone exacerbates obstructive renal injury by stimulating TNF-alpha production and increasing proapoptotic and profibrotic signaling. Am J Physiol Endocrinol Metab. 2008;294:E435–43.
Ridker PM, Rifai N, Pfeffer M, Sacks F, Lepage S, Braunwald E. Elevation of tumor necrosis factor-alpha and increased risk of recurrent coronary events after myocardial infarction. Circulation. 2000;101:2149–53.
Li D, Zhao L, Liu M, Du X, Ding W, Zhang J, et al. Kinetics of tumor necrosis factor alpha in plasma and the cardioprotective effect of a monoclonal antibody to tumor necrosis factor alpha in acute myocardial infarction. Am Heart J. 1999;137:1145–52.
Giannakopoulou M, Bozas E, Philippidis H, Stylianopoulou F. Protooncogene c-fos involvement in the molecular mechanism of rat brain sexual differentiation. Neuroendocrinology. 2001;73:387–96.
Zhang S, Zhang M, Goldstein S, Li Y, Ge J, He B, et al. The effect of c-fos on acute myocardial infarction and the significance of metoprolol intervention in a rat model. Cell Biochem Biophys. 2013;65:249–55.
US Food and Drug Administration, Center for Drug Evaluation and Research. FDA Drug Safety Communication: FDA cautions about using testosterone products for low testosterone due to aging; requires labeling change to inform of possible increased risk of heart attack and stroke with use. 2015. https://www.fda.gov/Drugs/DrugSafety/ucm436259.htm. Accessed 19 Apr 2018.
Vigen R, O’Donnell CI, Barón AE, Grunwald GK, Maddox TM, Bradley SM, et al. Association of testosterone therapy with mortality, myocardial infarction, and stroke in men with low testosterone levels. JAMA. 2013;310:1829–36.
Finkle WD, Greenland S, Ridgeway GK, Adams JL, Frasco MA, Cook MB, et al. Increased risk of non-fatal myocardial infarction following testosterone therapy prescription in men. PLoS One. 2014;9:e85805.
Morgentaler A, Zitzmann M, Traish AM, Fox AW, Jones TH, Maggi M, et al. Fundamental concepts regarding testosterone deficiency and treatment: international expert consensus resolutions. Mayo Clin Proc. 2016;91:881–96.
Ali WHAB. Ciprofloxacin-associated posterior reversible encephalopathy. BMJ Case Rep. 2013;2013. https://doi.org/10.1136/bcr-2013-008636.
Patel AS, Supan EM, Ali SN. Toxic epidermal necrolysis associated with rifaximin. Am J Health Syst Pharm. 2013;70:874–6.
Sharma D, Ivanovski S, Slevin M, Hamlet S, Pop TS, Brinzaniuc K, et al. Bisphosphonate-related osteonecrosis of jaw (BRONJ): diagnostic criteria and possible pathogenic mechanisms of an unexpected anti-angiogenic side effect. Vasc Cell. 2013;5:1.
Smalheiser NR, Swanson DR. Using ARROWSMITH: a computer-assisted approach to formulating and assessing scientific hypotheses. Comput Methods Programs Biomed. 1998;57:149–53.
Shang N, Xu H, Rindflesch TC, Cohen T. Identifying plausible adverse drug reactions using knowledge extracted from the literature. J Biomed Inform. 2014;52:293–310.
Hristovski D, Rindflesch T, Peterlin B. Using literature-based discovery to identify novel therapeutic approaches. Cardiovasc Hematol Agents Med Chem. 2013;11:14–24.
Yetisgen-Yildiz M, Pratt W. A new evaluation methodology for literature-based discovery systems. J Biomed Inform. 2009;42:633–43.
Preiss J, Stevenson M, Gaizauskas R. Exploring relation types for literature-based discovery. J Am Med Inform Assoc. 2015;22:987–92.
Tari L, Vo N, Liang S, Patel J, Baral C, Cai J. Identifying novel drug indications through automated reasoning. PLoS One. 2012;7:e40946.
Andronis C, Sharma A, Virvilis V, Deftereos S, Persidis A. Literature mining, ontologies and information visualization for drug repurposing. Brief Bioinform. 2011;12:357–68.
Doulaverakis C, Nikolaidis G, Kleontas A, Kompatsiaris I. Panacea, a semantic-enabled drug recommendations discovery framework. J Biomed Semant. 2014;5:13.
Acknowledgements
The authors would like to thank Mr Alexander Gartland for fruitful discussions that helped to improve the quality of the manuscript.
The MedDRA® trademark is owned by the International Federation of Pharmaceutical Manufacturers and Associations (IFPMA) on behalf of the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was supported in part by the National Institutes of Health (Grant 1U01CA207160).
Conflict of interest
Mary La, Alexander Sedykh, Denis Fourches, Eugene Muratov, and Alexander Tropsha declare no conflict of interests that are directly relevant to the content of this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
La, M.K., Sedykh, A., Fourches, D. et al. Predicting Adverse Drug Effects from Literature- and Database-Mined Assertions. Drug Saf 41, 1059–1072 (2018). https://doi.org/10.1007/s40264-018-0688-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40264-018-0688-5