Abstract
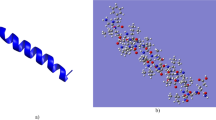
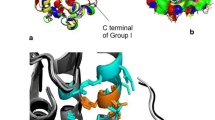
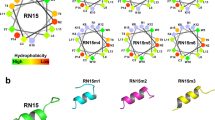
The emergence of antibiotic-resistant pathogens generates impairment to human health. U1-SCTRX-lg1a is a peptide isolated from a phospholipase D extracted from the spider venom of Loxosceles gaucho with antimicrobial activity against Gram-negative bacteria (between 1.15 and 4.6 μM). The aim of this study was to suggest potential receptors associated with the antimicrobial activity of U1-SCTRX-lg1a using in silico bioinformatics tools. The search for potential targets of U1-SCRTX-lg1a was performed using the PharmMapper server. Molecular docking between U1-SCRTX-lg1a and the receptor was performed using PatchDock software. The prediction of ligand sites for each receptor was conducted using the PDBSum server. Chimera 1.6 software was used to perform molecular dynamics simulations only for the best dock score receptor. In addition, U1-SCRTX-lg1a and native ligand interactions were compared using AutoDock Vina software. Finally, predicted interactions were compared with the ligand site previously described in the literature. The bioprospecting of U1-SCRTX-lg1a resulted in the identification of three hundred (300) diverse targets (Table S1), forty-nine (49) of which were intracellular proteins originating from Gram-negative microorganisms (Table S2). Docking results indicate Scores (10,702 to 6066), Areas (1498.70 to 728.40) and ACEs (417.90 to – 152.8) values. Among these, NAD + NH3-dependent synthetase (PDB ID: 1wxi) showed a dock score of 9742, area of 1223.6 and ACE of 38.38 in addition to presenting a Normalized Fit score of 8812 on PharmMapper server. Analysis of the interaction of ligands and receptors suggests that the peptide derived from brown spider venom can interact with residues SER48 and THR160. Furthermore, the C terminus (– 7.0 score) has greater affinity for the receptor than the N terminus (– 7.7 score). The molecular dynamics assay shown that free energy value for the protein complex of – 214,890.21 kJ/mol, whereas with rigid docking, this value was – 29.952.8 sugerindo that after the molecular dynamics simulation, the complex exhibits a more favorable energy value compared to the previous state. The in silico bioprospecting of receptors suggests that U1-SCRTX-lg1a may interfere with NAD + production in Escherichia coli, a Gram-negative bacterium, altering the homeostasis of the microorganism and impairing growth.



Similar content being viewed by others
Data and software availability
The physicochemical properties were determined via the Heliquest server https://heliquest.ipmc.cnrs.fr/. Potential receptors were screened through PharmMapper available in http://www.lilab-ecust.cn/pharmmapper/. Sequence and files of receptors were downloaded from website protein data bank https://www.rcsb.org/. A molecular docking method was used: PatchDock https://bioinfo3d.cs.tau.ac.il/PatchDock/ and https://vina.scripps.edu/. Ligand and receptor interaction, molecular presentation was built by free software UCSF chimera (version 1.16) https://www.cgl.ucsf.edu/chimera/. For prediction of ligand site, it was used by the PDBsum server http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/. All files used in this study are available in https://github.com/loxoscelesgaucho/loxosceles-in-silico.git.
References
Banerjee S et al (2003) Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation. Biochemistry 42(12):3446–3456. https://doi.org/10.1021/bi0206917
Benfield AH, Henriques ST (2020) Mode-of-action of antimicrobial peptides: membrane disruption vs intracellular mechanisms. Front Med Technol 2:610997. https://doi.org/10.3389/fmedt.2020.610997
Caycedo MI et al (2018) Antibiotic resistance: origins, evolution and healthcare-associated infections. Rev Salud Uninorte 34(2):494–505
Chan C et al (2004) Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci 101(49):17084–17089
Chaves-Moreira D et al (2017) Highlights in the knowledge of brown spider toxins. J Venom Anim Toxins Incl Trop Dis 23(1):1–12. https://doi.org/10.1186/s40409-017-0097-8
Chaves-Moreira D et al (2019) Brown spider (Loxosceles) venom toxins as potential biotools for the development of novel therapeutics. Toxins 11(6):355
da Silva BS et al (2023) Doderlin: Isolation and characterization of a broad-spectrum antimicrobial peptide from Lactobacillus acidophilus. Res Microbiol 174(3):103995
Dal Mas C et al (2019) Effects of the natural peptide crotamine from a South American rattlesnake on Candida auris, an emergent multidrug antifungal resistant human pathogen. Biomolecules 9(6):205
Duhovny D, Nussinov R, Wolfson HJ (2002) Efficient unbound docking of rigid molecules. International workshop on algorithms in bioinformatics. Springer, Berlin, Heidelberg, pp 185–200
Ferri M, Ranucci E, Romagnoli P, Giaccone V (2017) Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr 57(13):2857–2876
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the expasy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, pp 571–607
Gautier R et al (2008) HELIQUEST: A web server to screen sequences with specic α-helical properties. Bioinformatics 24:2101–2102
Glaab E (2016) Building a virtual ligand screening pipeline using free software: a survey. Brief Bioinform 17(2):352–366
Harris F, Dennison SR, Phoenix DA (2009) Anionic antimicrobial peptides from eukaryotic organisms. Curr Protein Pept Sci 10:585–606
Jauch R et al (2005) Structures of Escherichia coli NAD synthetase with substrates and products reveal mechanistic rearrangements. J Biol Chem 280(15):15131–15140
Landecker H (2015) Antibiotic resistance and the biology of history. Body Soc 22(4):19–52
Li X et al (2022) Antimicrobial mechanisms and clinical application prospects of antimicrobial peptides. Molecules 27(9):2675
Liu X et al (2010) PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res 38(suppl_2):W609–W614
Lowe RT (1832) Descriptions of two species of Araneidae, natives of Madeira. Zool J 5:320–326
Muniz Seif EJ, Icimoto MY, da Silva Junior PI (2023) In silico bioprospecting of receptors for Doderlin: an antimicrobial peptide isolated from Lactobacillus acidophilus. In Silico Pharmacol 11(1):11
Nelson DL, Cox MM (2022) Princípios de bioquímica de Lehninger. Aminoácidos, peptídeos e proteínas. Artmed Editora, pp 78–84
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Piontek K et al (1990) Structure determination and refinment of Bacillus stearothermophilus lactate dehydrogenase. Proteins Struct, Funct, Bioinform 7(1):74–92
Riciluca KCT et al (2012) Rondonin an antifungal peptide from spider (Acanthoscurria rondoniae) haemolymph. Res Immunol 2:66–71
Ruppert EE, Barnes RD (1996) Zoologia dos invertebrados, 6th edn. Roca, São Paulo
Schneidman-Duhovny D et al (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33(suppl_2):W363–W367
Segura-Ramírez PJ, Silva Júnior PI (2018) Loxosceles gaucho spider venom: an untapped source of antimicrobial agents. Toxins 10(12):522
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Wang X et al (2016) Enhancing the enrichment of pharmacophore-based target prediction for the polypharmacological profiles of drugs. J Chem Inf Model 56:1175–1183
Wang X et al (2017a) PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res 45:W356–W360
Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, Lai L, Pei J, Li H (2017b) Atualização do PharmMapper 2017: um servidor da Web para identificação potencial de alvos de drogas com um banco de dados abrangente de farmacoforos alvo. Res Ácidos Nucleicos 45:W356–W360
Welford RWD et al (2005) Incorporation of oxygen into the succinate co-product of iron (II) and 2-oxoglutarate dependent oxygenases from bacteria, plants and humans. FEBS Lett 579(23):5170–5174
Wilmouth RC et al (2002) Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure 10(1):93–103
Yang J, Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181
Zhang C, Freddolino PL, Zhang Y (2017) COFACTOR: improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res 45:W291-299
Zheng W, Zhang C, Li Y, Pearce R, Bell EW, Zhang Y (2021) Folding non-homology proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep Methods 1:100014
Acknowledgements
We thank all the team of the Protein Chemistry Laboratory at the Laboratory from Applied Toxinology (LETA—Butantan Institute, Brazil) for the constant support and encouragement.
Funding
This research received financial support from the Research Support Foundation of the State of São Paulo (FAPESP/CeTICS), grant number 2013/07467–1, Brazilian National Council for Scientific and Technological Development (CNPq), grant numbers 472744/2012–7 and 161722/2021–0, and from Higher Education Personnel Improvement Coordination (CAPES) process number 88887.663437/2022–00.
Author information
Authors and Affiliations
Contributions
Names in alphabetical order. Conceptualization and methodology: ASO, EJMS and P.I.S.Jr.; validation, formal analysis, investigation, resources, and data curation:ASO, EJMS and P.I.S.Jr; writing original draft: ASO and EJMS; review and editing, and visualization: ASO and EJMS and P.I.S.Jr.; supervision: P.I.S.Jr.; project administration and funding acquisition: P.I.S.Jr. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
de Oliveira, A.S., Muniz Seif, E.J. & da Silva Junior, P.I. In silico prospection of receptors associated with the biological activity of U1-SCTRX-lg1a: an antimicrobial peptide isolated from the venom of Loxosceles gaucho. In Silico Pharmacol. 12, 15 (2024). https://doi.org/10.1007/s40203-024-00190-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-024-00190-8