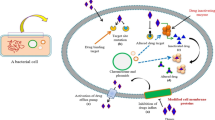
Abstract
The present study was designed to identify and analyze the targets of thymoquinone on drug resistant pathogens employing in silico tools. The target identification was performed using STITCH tool, followed by the functional analysis of protein targets by VICMPred. Further, VirulentPred was used to determine the nature of virulence of target proteins. The putative epitopes present on the virulent proteins were identified using BepiPred tool. The subcellular location of the virulent proteins was assessed using PSORTb. The results showed multiple targets of the pathogens being targeted. The nitric-oxide synthase-like protein of Staphylococcus aureus and acetyltransferase family protein, histone acetyltransferase HPA2, GNAT family acetyltransferase of Acinetobacter baumannii was found to be the virulent proteins interacting with thymoquinone. Molinspiration assessments showed zero violations suggesting the druggability of TQ. The study unveils the molecular mechanisms underlying the antimicrobial effect of thymoquinone as demonstrated by in silico procedures.




Similar content being viewed by others
References
Bakathir HA, Abbas NA (2011) Detection of the antibacterial effect of Nigella sativa ground seeds with water. Afr J Tradit Complement Altern Med 8(2):159–164
Chaieb K, Kouidhi B, Jrah H, Mahdouani K, Bakhrouf A (2011) Antibacterial activity of Thymoquinone, an active principle of Nigella sativa and its potency to prevent bacterial biofilm formation. BMC Complement Altern Med 11:1–6
Dönnes P, Annette H (2004) Predicting protein subcellular localization: past, present, and future. Genomics Proteomics Bioinform 2(4):209–215
Gardy JL, Brinkman F (2006) Methods for predicting bacterial protein subcellular localization. Nat Rev Microbiol 4:741–751
Garg A, Gupta D (2008) VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinform 9:62
Goyal SN, Prajapati CP, Gore PR, Patil CR, Mahajan UB, Sharma C, Talla SP, Ojha SK (2017) Therapeutic potential and pharmaceutical development of thymoquinone: a multi targeted molecule of natural origin. Front Pharmacol 8:656
Hanafy MS, Hatem ME (1991) Studies on the antimicrobial activity of Nigella sativa seed (black cumin). J Ethnopharmacol 34(2–3):275–278
Hannan A, Saleem S, Chaudhary S, Barkaat M, Arshad MU (2008) Anti-bacterial activity of Nigella sativa against clinical isolates of methicillin resistant Staphylococcus aureus. J Ayub Med Coll Abbottabad 20:72–74
Harzallah HJ, Kouidhi B, Flamini G, Bakhrouf A, Mahjoub T (2011) Chemical composition, antimicrobial potential against cariogenic bacteria and cytotoxic activity of Tunisian Nigella sativa essential oil and thymoquinone. Food Chem 129:1469–1474
Hasan NA, Nawahwi MZ, Malek HA (2013) Anti-microbial activity of Nigella sativa seed extract. Sains Malays 42:143–147
Hosseinzadeh H, Fazly Bazzaz BS, Haghi MM (2007) Antibacterial activity of total extracts and essential oil of Nigella sativa .L seeds in mice. Pharmacol Online 2:429–435
Jespersen MC, Peters B, Nielsen M (2017) BepiPred-2.0: improving sequence based B cell epitope prediction using conformational epitopes. Nucleic Acids Res 45(W1):W24–W29
Khan AR, Kour K (2016) Wide spectrum antibacterial activity of Nigella sativa L. seeds. IOSR J Pharm 6(7):12–16
Kiari FZ, Meddah B, Meddah ATT (2018) In vitro study on the activity of essential oil and methanolic extract from Algerian Nigella sativa L. Seeds on the growth kinetics of micro-organisms isolated from the buccal cavities of periodontal patients. Saudi Dent J 30(4):312–323
Klevens RM, Edwards JR, Richards CL (2007) Estimating health care-associated infections and deaths in U.S. Hospitals, 2002. Public Health Rep 122(2):160–166
Larsen JE, Lund O, Nielsen M (2006) Improved method for predicting linear B-cell epitopes. Immunome Res 24:2
Navratna V, Reddy G, Gopal B (2015) Structural basis for the catalytic mechanism of homoserine dehydrogenase. Acta Crystallogr D Biol Crystallogr 71:1216–1225
Pendleton JN, Gorman SP, Gilmore BF (2013) Clinical relevance of the ESKAPE pathogens. Expert Rev Antiinfective Ther 11(3):297–308
Rice LB (2010) Progress and challenges in implementing the research on ESKAPE pathogens. Infect Control Hosp Epidemiol 31(1):S7–S10
Ryan A, Kaplan E, Nebel J-C, Polycarpou E, Crescente V, Lowe E, Preston GM, Sim E (2014) Identification of NAD(P)H quinone oxidoreductase activity in azoreductases from P. aeruginosa: azoreductases and NAD(P)H quinone oxidoreductases belong to the same FMN-dependent superfamily of enzymes. PLoS One 10:9(6):e98551
Saha S, Raghava GPS (2006) VICMpred: SVM-based method for the prediction of functional proteins of gram-negative bacteria using amino acid patterns and composition. Genom Proteom Bioinform 4:42–47
Salih B, Sipahi T, Donmez EO (2009) Ancient nigella seeds from Boyali Hoyuk in north-central Turkey. J Ethnopharmacol 124:416–420
Santajit S, Indrawattana N (2016) Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res Int 2475067:8 pages
Scibior D, Czeczot H (2006) Catalase: structure, properties, functions. Postepy Hig Med Dosw 60:170–180
Szklarczyk D, Santos A, Von Mering C et al (2016) STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acid Res 44:D380–D384
Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL (2018) Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327
Tavolieri AM, Murray DT, Askenasy I, Pennington JM, McGarry L, Stanley CB, Stroupe ME (2019) NADPH-dependent sulfite reductase flavoprotein adopts an extended conformation unique to this diflavin reductase. J Struct Biol 205(2):170–179
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31(2):455–461
Tomar JS, Peddinti RK (2017) A. Baumannii histone acetyl transferase Hpa2: optimization of homology modeling, analysis of protein-protein interaction and virtual screening. J Biomol Struct Dyn 35(5):1115–1126
Ugur AR, Dagi HT, Ozturk B, Tekin G, Findik D (2016) Assessment of in vitro antibacterial activity and cytotoxicity effect of Nigella sativa oil. Pharmacogn Mag 12(4):S471–S474
Verbanac D, Jeli D, Stepanik V et al (2005) Combined in silico and in vitro approach to drug screening. Croat Chem Acta 78(2):133–139
Weininger D (1988) SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci. 28(1):31–36
Yu NK, Wagner JR, Laird MR (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O et al (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644
Zhao Y, Xi Q, Xu Q, He M, Ding J, Dai Y, Keller NP, Zheng W (2015) Correlation of nitric oxide produced by an inducible nitric oxide synthase-like protein with enhanced expression of the phenylpropanoid pathway in Inonotus obliquus co-cultured with Phellinus morii. Appl Microbiol Biotechnol 99(10):4361–4372
Acknowledgements
None to declare.
Funding
No grant or financial support received for the study.
Author information
Authors and Affiliations
Contributions
ASSG made substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data; drafted the work or revised it critically for important intellectual content; approved the version to be published; and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. SG performed the docking analysis and interpretation with final validation of the manuscript. AP approved the version to be published; and 4) agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. JVP substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data; drafting and final validation of the manuscript and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Girija, A.S.S., Gnanendra, S., Paramasivam, A. et al. Delineating the potential targets of thymoquinone in ESKAPE pathogens using a computational approach. In Silico Pharmacol. 9, 52 (2021). https://doi.org/10.1007/s40203-021-00111-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-021-00111-z