Abstract
Purpose
Escherichia coli O157:H7 is one of the major foodborne pathogens of global public concern. Bacteriophages (phages) have emerged as a promising alternative to antibiotics for controlling pathogenic bacteria. Here, a lytic E. coli O157:H7-specific phage (KFS-EC) was isolated, identified, and characterized to evaluate its potential as a biocontrol agent for E. coli O157:H7.
Methods
KFS-EC was isolated from slaughterhouse in Korea. Morphological analysis, genomic analysis and several physiological tests were performed to identify and characterize the KFS-EC.
Results
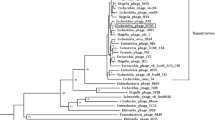
A specificity test indicated KFS-EC was strictly specific to E. coli O157:H7 strains among 60 bacterial strains tested. Morphological and phylogenetic analyses confirmed that KFS-EC belongs to the Rb49virus genus, Tevenvirinae subfamily, and the Myoviridae family of phages. KFS-EC genome consists of 164,725 bp and a total of 270 coding sequence features, of which 114 open reading frames (ORFs) were identified as phage functional genes. KFS-EC does not contain genes encoding lysogenic property and pathogenicity, which ensure its safe application. KFS-EC was relatively stable (~1 log decrease) under stressed conditions such as temperatures (20 °C–50 °C), pHs (3–11), organic solvents (ethanol and chloroform), and biocides (0.1% citric acid, 1% citric acid, and 0.1% peracetic acid). KFS-EC was able to inhibit E. coli O157:H7 efficiently at a multiplicity of infection (MOI) of 0.01 for 8 h with greater inhibitory effect and durability and was stable at 4 °C and 22 °C over a 12-week storage period.
Conclusions
Our results suggest that KFS-EC could be used as a biocontrol agent to E. coli O157:H7.







Similar content being viewed by others
References
Adamu MT, Shamsul BMT, Desa MN, Khairani-Bejo S. A review on Escherichia coli O157:H7-the super pathogen. Health Environ J. 2014;5:78–93.
Saxena T, Kaushik P, Mohan MK. Prevalence of E. coli O157:H7 in water sources: an overview on associated diseases, outbreaks and detection methods. Diagn Microbiol Infect Dis. 2015;82:249–64.
Saeedi P, Yazdanparast M, Behzadi E, Salmanian AH, Mousavi SL, Nazarian S, et al. A review on strategies for decreasing E. coli O157:H7 risk in animals. Microb Pathog. 2017;103:186–95.
Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–5.
Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40.
Majowicz SE, Scallan E, Jones-Bitton A, Sargeant JM, Stapleton J, Angulo FJ, et al. Global incidence of human Shiga toxin–producing Escherichia coli infections and deaths: a systematic review and knowledge synthesis. Foodborne Pathog Dis. 2014;11:447–55.
Holvoet K, Sampers I, Callens B, Dewulf J, Uyttendaele M. Moderate prevalence of antimicrobial resistance in Escherichia coli isolates from lettuce, irrigation water, and soil. Appl Environ Microbiol. 2013;79:6677–83.
Schroeder CM, Zhao C, DebRoy C, Torcolini J, Zhao S, White DG, et al. Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Appl Environ Microbiol. 2002;68:576–81.
Francino MP. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol. 2016;6:1543.
Choi IY, Lee J-H, Kim H-J, Park M-K. Isolation and characterization of a novel broad-host-range bacteriophage infecting Salmonella enterica subsp. enterica for biocontrol and rapid detection. J Microbiol Biotechnol. 2017;27:2151–5.
Amarillas L, Chaidez C, González-Robles A, Lugo-Melchor Y, León-Félix J. Characterization of novel bacteriophage phiC119 capable of lysing multidrug-resistant Shiga toxin-producing Escherichia coli O157:H7. PeerJ. 2016;4:e2423.
Oh J-H, Park M-K. Recent trends in Salmonella outbreaks and emerging technology for biocontrol of Salmonella using phages in foods: a review. J Microbiol Biotechnol. 2017;27:2075–88.
Lee H, Ku H-J, Lee D-H, Kim Y-T, Shin H, Ryu S, et al. Characterization and genomic study of the novel bacteriophage HY01 infecting both Escherichia coli O157:H7 and Shigella flexneri: potential as a biocontrol agent in food. PLoS One. 2016;11:e0168985.
Hudson JA, Billington C, Carey-Smith G, Greening G. Bacteriophages as biocontrol agents in food. J Food Prot. 2005;68:426–37.
Carlton RM. Phage therapy: past history and future prospects. Arch Immunol Ther Exp. 1999;47:267–74.
Ackermann H-W, DuBow M, Jarvis A, Jones L, Krylov V, Maniloff J, et al. The species concept and its application to tailed phages. Arch Virol. 1992;124:69–82.
Mirzaei MK, Nilsson AS. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One. 2015;10:e0118557.
Bushnell, B. BB Map 2014. https://sourceforge.net/projects/bbmap/ Accessed 14 Feb 2019.
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75.
Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132.
Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21.
Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44:D694–7.
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4.
McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–57.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9.
Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Res. 2017;46:D708–17.
Meier-Kolthoff JP, Göker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33:3396–404.
Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60.
Lefort V, Desper R, Gascuel O. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol. 2015;32:2798–800.
Farris JS. Formal definitions of paraphyly and polyphyly. Syst Zool. 1974;23:548–54.
Rambaut A. FigTree-version 1.4.3, a graphical viewer of phylogenetic trees; 2017. http://tree.bio.ed.ac.uk/software/figtree/ Accessed 14 Feb 2019.
Göker M, García-Blázquez G, Voglmayr H, Tellería MT, Martín MP. Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One. 2009;4:e6319.
Meier-Kolthoff JP, Hahnke RL, Petersen J, Scheuner C, Michael V, Fiebig A, et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 2014;9:2.
Ackermann H-W. 5500 phages examined in the electron microscope. Arch Virol. 2007;152:227–43.
Hudson JA, Billington C, Cornelius AJ, Wilson T, On SLW, Premaratne A, et al. Use of a bacteriophage to inactivate Escherichia coli O157:H7 on beef. Food Microbiol. 2013;36:14–21.
Raya RR, Varey P, Oot RA, Dyen MR, Callaway TR, Edrington TS, et al. Isolation and characterization of a new T-even bacteriophage, CEV1, and determination of its potential to reduce Escherichia coli O157:H7 levels in sheep. Appl Environ Microbiol. 2006;72:6405–10.
Park M, Lee J-H, Shin H, Kim M, Choi J, Kang D-H, et al. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157: H7. Appl Environ Microbiol. 2012;78:58–69.
Denou E, Bruttin A, Barretto C, Ngom-Bru C, Brüssow H, Zuber S. T4 phages against Escherichia coli diarrhea: potential and problems. Virology. 2009;388:21–30.
Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A. 2011;108:20142–7.
Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol J. 2010;7:292.
Riede I, Drexler K, Schwarz H, Henning U. T-even-type bacteriophages use an adhesin for recognition of cellular receptors. J Mol Biol. 1987;194:23–30.
Hamdi S, Rousseau GM, Labrie SJ, Tremblay DM, Kourda RS, Slama KB, et al. Characterization of two polyvalent phages infecting Enterobacteriaceae. Sci Rep. 2017;7:40349.
Song JY, Yoo RH, Jang SY, Seong W-K, Kim S-Y, Jeong H, et al. Genome sequence of enterohemorrhagic Escherichia coli NCCP15658. J Bacteriol. 2012;194:3749–50.
Trojet SN, Caumont-Sarcos A, Perrody E, Comeau AM, Krisch HM. The gp38 adhesins of the T4 superfamily: a complex modular determinant of the phage’s host specificity. Genome Biol Evol. 2011;3:674–86.
Adam E, Groenenboom AE, Kurm V, Rajewska M, Schmidt R, Tyc O, et al. Controlling the microbiome: microhabitat adjustments for successful biocontrol strategies in soil and human gut. Front Microbiol. 2016;7:1079.
Loc-Carrillo C, Abedon ST. Pros and cons of phage therapy. Bacteriophage. 2011;1:111–4.
Amarillas L, Rubí-Rangel L, Chaidez C, González-Robles A, Lightbourn-Rojas L, León-Félix J. Isolation and characterization of phiLLS, a novel phage with potential biocontrol agent against multidrug-resistant Escherichia coli. Front Microbiol. 2017;8:1355.
Meaden S, Koskella B. Exploring the risks of phage application in the environment. Front Microbiol. 2013;4:358.
Wang J, Niu YD, Chen J, Anany H, Ackermann H-W, Johnson RP, et al. Feces of feedlot cattle contain a diversity of bacteriophages that lyse non-O157 Shiga toxin-producing Escherichia coli. Can J Microbiol. 2015;61:467–75.
Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, et al. Complete genome sequence of enterohemorrhagic Eschelichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22.
Weiserová M, Ryu J. Characterization of a restriction modification system from the commensal Escherichia coli strain A0 34/86 (O83:K24:H31). BMC Microbiol. 2008;8:106.
Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:86–156.
Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27.
Verma V, Harjai K, Chhibber S. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: a potential therapeutic agent. Curr Microbiol. 2009;59:274–81.
Suarez VB, Reinheimer JA. Effectiveness of thermal treatments and biocides in the inactivation of Argentinian Lactococcus lactis phages. J Food Prot. 2002;65:1756–9.
Tomat D, Balagué C, Aquili V, Verdini R, Quiberoni A. Resistance of phages lytic to pathogenic Escherichia coli to sanitisers used by the food industry and in home settings. Int J Food Sci Technol. 2018;53:533–40.
Xu Y, Yu X, Gu Y, Huang X, Liu G, Liu X. Characterization and genomic study of phage Vb_EcoS-B2 infecting multidrug-resistant Escherichia coli. Front Microbial. 2018;9:793.
Peng Q, Yuan Y. Characterization of a newly isolated phage infecting pathogenic Escherichia coli and analysis of its mosaic structural genes. Sci Rep. 2018;8:8086.
Kim M, Ryu S. Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl Environ Microbiol. 2011;77:2042–50.
Clark WA. Comparison of several methods for preserving bacteriophages. Appl Microbiol. 1962;10:466–71.
Jończyk E, Kłak M, Międzybrodzki R, Górski A. The influence of external factors on bacteriophages-review. Folia Microbiol. 2011;56:191–200.
Funding
This research was funded by the Basic Research Program through the National Research Foundation (NRF) of Korea founded by the Ministry of Education (NRF-2017R1D1A1B03035195).
Author information
Authors and Affiliations
Contributions
CL, IYC, DHP, and M-KP conducted this study, and drafted the manuscript. M-KP supervised the study in all steps. CL, IYC, and M-KP edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 157 kb)
Rights and permissions
About this article
Cite this article
Lee, C., Choi, I.Y., Park, D.H. et al. Isolation and characterization of a novel Escherichia coli O157:H7-specific phage as a biocontrol agent. J Environ Health Sci Engineer 18, 189–199 (2020). https://doi.org/10.1007/s40201-020-00452-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40201-020-00452-5