Abstract
Although Escherichia coli is the most widely studied bacterial model organism and often considered to be the model bacterium per se, its type strain was until now forgotten from microbial genomics. As a part of the G enomic E ncyclopedia of B acteria and A rchaea project, we here describe the features of E. coli DSM 30083T together with its genome sequence and annotation as well as novel aspects of its phenotype. The 5,038,133 bp containing genome sequence includes 4,762 protein-coding genes and 175 RNA genes as well as a single plasmid. Affiliation of a set of 250 genome-sequenced E. coli strains, Shigella and outgroup strains to the type strain of E. coli was investigated using digital DNA:DNA-hybridization (dDDH) similarities and differences in genomic G+C content. As in the majority of previous studies, results show Shigella spp. embedded within E. coli and in most cases forming a single subgroup of it. Phylogenomic trees also recover the proposed E. coli phylotypes as monophyla with minor exceptions and place DSM 30083T in phylotype B2 with E. coli S88 as its closest neighbor. The widely used lab strain K-12 is not only genomically but also physiologically strongly different from the type strain. The phylotypes do not express a uniform level of character divergence as measured using dDDH, however, thus an alternative arrangement is proposed and discussed in the context of bacterial subspecies. Analyses of the genome sequences of a large number of E. coli strains and of strains from > 100 other bacterial genera indicate a value of 79-80% dDDH as the most promising threshold for delineating subspecies, which in turn suggests the presence of five subspecies within E. coli.
Similar content being viewed by others
Introduction
Despite more than 35,000 completed and ongoing bacterial genome-sequencing projects (including over 2,500 genomes from strains of the genus Escherichia) [1] and the fundamental importance of type strains for microbial taxonomy and nomenclature [2], the type strain of Escherichia coli, U5/41T, the most widely studied bacterial model organism and model bacterium per se, was until now neglected in microbial genomics; although strain K-12 substrain MG1665 was in 1997 the subject of one of the first ever published complete genome sequences [3]. By sequencing the genome of DSM 30083T, DSMZ’s culture of U5/41T, in the context of the Genomic Encyclopedia of Bacteria and Archaea [4], we filled this gap enabling not only the use of this strain as a taxonomic reference in genome sequence-based studies, but also providing access to novel data of an exciting organism whose phenotypic features differ in many ways from those of the often used E. coli lab strain K-12.
The first report on strains of the genus Escherichia (at that time termed “Bacterium coli commune”) were published in 1886 by Theodor Escherich [5] in the context of his professorial dissertation at University of Munich. Later in 1919, Castellani and Chalmers proposed the name Escherichia coli (E.sche.ri’chi.a, M.L. fem.n., Escherichia, in honor of Theodor Escherich; co’li, Gr.n. colon large intestine, colon, M.L. gen.n. coli of the colon) as the name for the type species of the genus Escherichia, which was accepted by the Judical Commission of the ICSB in 1958 [6] and included in the Approved Lists of Bacterial Names in 1980 [7].
Despite its enormous importance for microbiology and mostly due to a lack of type culture collections until the early 1920s, the original type cultures of E. coli got lost (just like those of the early isolates of other bacterial species). Strain U5/41T (= DSM 30083T = ATCC 11775T = WDCM 00090T) was isolated by Fritz Kauffmann at the State Serum Institute Copenhagen, Denmark in 1941 [8], from the urine of a patient with cystitis, and was accepted as neotype of E. coli in 1963 [9]. Figure 1 shows the original record card issued by the Danish State Serum Institute in Copenhagen for the deposit of U5/41T. Since then, E. coli DSM 30083T was a reference strain for many tests and applications, such as serotyping with the method of Ørskov and Ørskov [10], antimicrobial assays [11], ribotyping and multi-locus sequence typing [12], and the PCR amplification of the β-d-glucuronidase gene fragment (uidA) as tracer for fecal pollution in all kinds of waters [13]. As a model organism for genetics, biochemistry, metabolic reconstruction and pathway inference, genomics and metabolics of E. coli are well-studied topics, starting with the 1997 publication of the K-12 genome [3]. The reader is referred to studies of E. coli such as metabolic engineering for the production of chemicals and biofuels [14, 15], recombinant protein expression [16], the process of binary fission [17], DNA replication and segregation [18], small RNA regulators [19], genetics of the capsular machinery gene cluster [20], as well as comparative genomics [21] and the current status and the progress in clinically relevant E. coli strains [22, 23].
In this study we analyzed the genome sequence of E. coli DSM 30083T. We present a description of the genome sequencing and annotation and a summary classification together with a set of features for strain DSM 30083T, including novel aspects of its phenotype. Since only the availability of the type-strain genome allows for the application of state-of-the-art genome-based taxonomic methods, species affiliation of all strains with respect to the type strain was determined via digital DNA:DNA-hybridization (dDDH) similarities as computed by the Genome-to-Genome Distance Calculator [24], version 2 [25], and by evaluating the differences in genomic G+C content [26]. Phylogenomic analyses [24, 25] elucidate the evolutionary relationships between 250 E. coli strains, Shigella spp. and outgroup strains as well as the grouping within E. coli. The availability of the type-strain genome allows not only for assessing whether published genome sequences are actually from strains of E. coli but also for a potential division of E. coli into subspecies.
Organism features
Classification and features
16S rRNA gene analysis
The sequences of the seven 16S rRNA gene copies in the genome of DSM 30083T differ from each other by up to eleven nucleotides, and differ by up to ten nucleotides from the previously published 16S rRNA gene sequence (X80725), which contains three ambiguous base calls. The phylogenetic neighborhood of E. coli in a 16S rRNA gene-based tree inferred as previously described [27] is shown in Additional file 1.
The single genomic 16S rRNA gene sequence of E. coli DSM 30083T was compared with the Greengenes database for determining the weighted relative frequencies of taxa and (truncated) keywords as previously described [27]. The most frequently occurring genera were Escherichia (87.0%) and Shigella (13.0%) (131 hits in total). Regarding the 109 hits to sequences from representatives of the species, the average identity within HSPs was 99.8%, whereas the average coverage by HSPs was 100.0%. Regarding the five hits to sequences from other representatives of the genus, the average identity within HSPs was also 99.8%, whereas the average coverage by HSPs was 100.0%. Among all other species, the one yielding the highest score was Shigella flexneri (HQ407229), which corresponded to an identity of 99.9% and an HSP coverage of 100.0%. (Note that the Greengenes database uses the INSDC (=EMBL/NCBI/DDBJ) annotation, which is not an authoritative source for nomenclature or classification.) The highest-scoring environmental sequence was EF603461 (Greengenes short name ‘Salmonella typhimurium Exploits Inflammation Compete Intestinal Microbiota mouse cecum clone 16saw29-1c11.q1k’), which showed an identity of 99.9% and an HSP coverage of 100.0%. The most frequently occurring keywords within the labels of all environmental samples which yielded hits were ‘intestin’ (9.9%), ‘mous’ (6.1%), ‘inflamm’ (5.8%), ‘microbiota’ (5.7%) and ‘cecum, compet, exploit, salmonella, typhimurium’ (5.6%) (119 hits in total). The most frequently occurring keywords within the labels of those environmental samples which yielded hits of a higher score than the highest scoring species were ‘microbiota’ (12.5%), ‘cecum, compet, exploit, inflamm, intestin, mous, salmonella, typhimurium’ (10.0%) and ‘gut, lusitanicu, thorect’ (2.5%) (5 hits in total). These keywords fit well to the known ecology of E. coli.
Morphology and physiology
As described for the genus Escherichia, cells are Gram-negative, medium to long rods (Figure 2 and Table 1), motile by the means of peritrichous flagella, non-pigmented, chemo-organotrophic, oxidase-negative, facultative anaerobes. They produce acid and gas while fermenting d-glucose, lactose or other carbohydrates [28]. E. coli strains are able to grow at temperatures between 10°C and 45°C, with an optimum between 37°C and 42°C, and at pH 5.5-8.0 [28, 29]. Koser [30] showed that “Bacterium coli communis” utilizes propionic acid, n-butyric acid, succinic acid, malic acid, lactic acid and mucic acid as sole carbon sources, but neither citric acid, salts of citric acid, n- or iso-valeric acid, n-caprionic acid, tartaric acid, oxalic acid, benzoic acid, salicylic acid nor o-phthalic acid. Based on the description by Kauffmann (Figure 1), strain DSM 30083T grows on d-trehalose, d-sorbitol, d-mannitol, l-rhamnose, d-glucose, d-maltose, α-d-lactose, d-arabinose, but does not grow on dulcitol, d-xylose, sucrose, adonitol, citric acid, inositol and gelatin and growth varies on d-salicin. Strain DSM 30083T belongs to E. coli “var. communis” (representatives were mostly isolated from feces), because the strain does not ferment sucrose or salicin [31]. Strain DSM 30083T is able to ferment lactose (Figure 1), which is a characteristic criterion for the differentiation against representatives of Shigella and Salmonella [28, 29]. Comparable to most strains of E. coli, strain DSM 30083T is positive for indole production, nitrate reduction, and urease but hydrogen-sulfide negative (Figure 1) [28]. Additionally, Huys et al. [32] described strain ATCC 11775T as being positive for d-raffinose and acetate utilization, positive for lysin-decarboxylase and ornithine-decarboxylase activity, and negative for growth on d-arabitol, d-cellobiose and in the presence of KCN. Furthermore, E. coli utilizes mucic acid, does not produce acetoin (Voges–Proskauer negative), and does not utilize malonate [29].
We used phenotyping with the OmniLog instrument [Biolog Inc., Hayward, CA] to elucidate whether or not strain DSM 30083T might be able to utilize further substrates. A comparison of E. coli DSM 30083T and E. coli DSM 18039 (a K-12 MG1655 derivative with almost K-12 wild-type features) with Generation-III microplates run in an OmniLog phenotyping instrument was conducted by Vaas et al. [44]. These data also serve as exemplars for the substrate-information and feature-selection facilities in the tutorial of the opm package [45] for analyzing phenotype microarray data in the R statistical environment [46]. As shown in that tutorial, among the substrates contained in Generation-II plates, carbohydrates make the main difference between the two strains, with DSM 30083T mostly reacting more strongly than DSM 18039.
The utilization of carbon compounds by E. coli DSM 30083T grown at 37°C in LB medium (DSMZ medium no. 381) [41] was also determined for this study using PM-01 and PM-02 microplates [Biolog Inc., Hayward, CA]. These plates were inoculated at 37°C with dye A and a cell suspension at a cell density of 85% turbidity. The exported measurement data were further analyzed with opm using its functionality for statistically estimating parameters from the respiration curves such as the maximum height, and automatically translating these values into negative, ambiguous, and positive reactions. The reactions were recorded in two individual biological replicates, and results that differed between the two replicates were regarded as ambiguous.
On PM-01 microplates, DSM 30083T was positive for l-arabinose, N-acetyl-d-glucosamine, d-saccharic acid, succinic acid, d-galactose, l-aspartic acid, l-proline, d-alanine, d-trehalose, d-mannose, d-serine, d-sorbitol, glycerol, l-fucose, d-glucuronic acid, d-gluconic acid, d,l-α-glycerol-phosphate, l-lactic acid, d-mannitol, l-glutamic acid, d-glucose-6-phosphate, d-galactonic acid-γ-lactone, d,l-malic acid, d-ribose, tween 20, l-rhamnose, d-fructose, acetic acid, d-glucose, d-maltose, d-melibiose, thymidine, l-asparagine, d-glucosaminic acid, tween 40, α-keto-glutaric acid, α-methyl-d-galactoside, α-d-lactose, lactulose, uridine, l-glutamine, α-d-glucose-1-phosphate, d-fructose-6-phosphate, β-methyl-d-glucoside, maltotriose, 2′-deoxy-adenosine, adenosine, gly-asp, fumaric acid, bromo-succinic acid, propionic acid, glycolic acid, glyoxylic acid, inosine, gly-glu, l-serine, l-threonine, l-alanine, ala-gly, N-acetyl-β-d-mannosamine, mono-methyl succinate, methyl pyruvate, d-malic acid, l-malic acid, gly-pro, l-lyxose, glucuronamide, pyruvic acid, l-galactonic acid-γ-lactone and d-galacturonic acid.
The strain was negative for the negative control, dulcitol, d-xylose, d-aspartic acid, α-keto-butyric acid, sucrose, m-tartaric acid, tween 80, α-hydroxy-glutaric acid-γ-lactone, α-hydroxy-butyric acid, adonitol, citric acid, myo-inositol, d-threonine, mucic acid, d-cellobiose, tricarballylic acid, acetoacetic acid, p-hydroxy-phenylacetic acid, m-hydroxy-phenylacetic acid, tyramine, d-psicose, β-phenylethylamine and ethanolamine.
Ambiguous results were obtained with sodium formate and 1,2-propanediol.
On PM-02 microplates, DSM 30083T was positive for dextrin, N-acetyl-d-galactosamine, N-acetyl-neuraminic acid, β-d-allose, d-arabinose, 3-O-β-d-galactopyranosyl-d-arabinose, d-lactitol, β-methyl-d-galactoside, β-methyl-d-glucuronic acid, d-raffinose, l-sorbose, d-tagatose, d-glucosamine, β-hydroxy-butyric acid, d-lactic acid methyl ester, melibionic acid, l-alaninamide and dihydroxy-acetone.
The strain was negative for the negative control, chondroitin sulfate C, α-cyclodextrin, β-cyclodextrin, γ-cyclodextrin, gelatin, glycogen, inulin, laminarin, mannan, pectin, amygdalin, d-arabitol, l-arabitol, arbutin, 2-deoxy-d-ribose, m-erythritol, d-fucose, β-gentiobiose, l-glucose, d-melezitose, maltitol, α-methyl-d-glucoside, 3-O-methyl-d-glucose, α-methyl-d-mannoside, β-methyl-d-xylopyranoside, palatinose, d-salicin, sedoheptulosan, stachyose, turanose, xylitol, N-acetyl-d-glucosaminitol, γ-amino-n-butyric acid, δ-amino-valeric acid, butyric acid, capric acid, caproic acid, citraconic acid, d-citramalic acid, 2-hydroxy-benzoic acid, 4-hydroxy-benzoic acid, γ-hydroxy-butyric acid, α-keto-valeric acid, itaconic acid, 5-keto-d-gluconic acid, malonic acid, oxalic acid, oxalomalic acid, quinic acid, d-ribono-1,4-lactone, sebacic acid, sorbic acid, succinamic acid, d-tartaric acid, l-tartaric acid, acetamide, N-acetyl-l-glutamic acid, l-arginine, glycine, l-histidine, l-homoserine, l-hydroxyproline, l-isoleucine, l-leucine, l-lysine, l-methionine, l-ornithine, l-phenylalanine, l-pyroglutamic acid, l-valine, d,l-carnitine, butylamine (sec), d,l-octopamine, putrescine, 2,3-butanediol, 2,3-butanedione and 3-hydroxy-2-butanone. Ambiguous results were not observed on PM-02 microplates.
Results of the OmniLog phenotyping in PM-01 and PM-02 microplates (see Additional file 1 for further information) were in full agreement with growth experiments as described in the aforementioned literature with the sole exception of mucic acid [29], which was not metabolized by strain DSM 30083T in OmniLog phenotyping, at least not within the applied running time. In brief, strain DSM 30083T grows on succinic acid, d-sorbitol, l-lactic acid, d-mannitol, l-rhamnose, acetic acid, d-glucose, d-maltose, α-d-lactose, propionic acid, d-trehalose, d-malic acid, l-malic acid, d-arabinose, and d-raffinose, but does not grow on dulcitol, d-xylose, sucrose, m-tartaric acid, adonitol, citric acid, myo-inositol, d-cellobiose, gelatin, d-arabitol, d-salicin, butyric acid, malonic acid, oxalic acid, d-tartaric acid, and l-tartaric acid. Strain DSM 30083T grows on d-galacturonic acid, d-glucuronic acid, α-keto-glutaric acid and glutamic acid, which suggests a catabolism of d-glucuronic acid and d-galacturonic acid to α-keto-glutaric acid and further to glutamic acid via the mucic-acid pathway [47, 48].
We tested growth on further substrates by incubating strain DSM 30083T either on DSMZ medium 382 (M9) without glucose [41], supplemented with 20 mM substrate at 37°C for 72 h, or with API 20E strips (bioMérieux, Nürtingen, Germany) at 37°C. On API 20E strips (see Additional file 1) strain DSM 30083T was positive for β-galactosidase, l-lysine, l-ornithine, indole production, d-glucose, d-mannitol, d-sorbitol, l-rhamnose, d-melibiose, and l-arabinose, but negative for l-arginine, citrate, sulfide production, urease, l-tryptophane, acetoin production, gelatin, inositol, sucrose, amygdaline, and oxidase. In medium M9 strain DSM 30083T showed growth on l-glutamic acid, tween 20, N- acetyl-d-galactosamine, l-sorbose, and d-melibiose, but not on 1,2-propanediol, dulcitol, d-xylose, m- tartaric acid, and α-keto-butyric acid. In experiments conducted at DSMZ, strain DSM 30083T formed blue colonies on OXOID Brilliance ESBL Agar (P05302A, OXOID, UK) and utilized d-galactose and thus is both galactosidase- and glucuronidase-positive. Indicated by the positive result of pyruvic acid in the OmniLog phenotyping and the negative Voges–Proskauer test, strain DSM 30083T is able to utilize pyruvate but does not produce acetoin, a carbon storage and an intermediate to avoid acidification during fermentation [49].
Chemotaxonomy
To the best of our knowledge, data on the fatty acids or polar lipids of E. coli DSM 30083T are not available in the literature.
For details on the extensively studied molecular structure and chemical composition of the E. coli cell wall the reader is referred to Scheutz and Strockbine [29] and the literature listed therein. In brief, E. coli has a single peptidoglycan layer within the periplasm, consisting of n-acetylglucosamine and n-acetylmuramic acid linked to the tetrapeptide l-alanine, d-glutamic acid, meso-diaminopimelic acid and d-alanine. The outer membrane is a lipopolysaccharide layer consisting of (i) lipid A, (ii) the core region of the phosphorylated nonrepeating oligosaccharides, and (iii) the O-antigen polymer [28, 29].
E. coli, Shigella ssp. and Salmonella ssp. strains display a huge variety of lipopolysaccharide layer heat-stable somatic (O), capsular (K; “Kapsel”, the German word for capsule), flagellar filament (H), and fimbriae (F) antigens, which serve since a long time as the basis for serotyping [29]. K antigens are further subdivided into the L, B, and A categories, based on their physical properties [29]. The serotype of E. coli DSM 30083T is O1:K1(L1):H7.
Representatives of E. coli, as Gram-negative bacteria, are described to be intrinsically resistant to hydrophobic antibiotics (e.g. macrolites, novoviocins, rifamycins, actinomycin D, fusidic acid) and may have acquired further antibiotic resistances (e.g. aminoglycosides, β-lactam, chloramphenicol, sulfonamides, tetracyclines) [29]. We tested the antibiotic resistance of E. coli DSM 30083T on Müller-Hinton agar at 30°C. Strain DSM 30083T was resistant against the cell-envelope antibiotics bacitracin, oxacillin, penicillin G, teicoplanin and vancomycin as well as against the protein-synthesis inhibitors (50S subunit) clindamycin, lincomycin, linezolid, nystatin (antifungal) and quinupristin/dalfooristin. In contrast, strain DSM 30083T was susceptible to the cell-envelope antibiotics ampicillin, azlocillin, aztreonam, cefalotin, cefazolin, cefotaxime, ceftriaxone, colistin, fosfomycin, imipenem, mezlocillin, piperacillin/tazobactam, ticarcillin and polymyxin B, the protein-synthesis inhibitors (30S subunit) amikacin, doxycyclin, gentamicin, kanamycin, neomycin and tetracyclin, the protein-synthesis inhibitors (50S subunit) chloramphenicol and erythromycin as well as against the nucleic-acid inhibitors moxifloxacin, nitrofurantoin, norflaxacin, oflaxacin and pipemidic acid.
As reported by F. Kauffmann (Figure 1) and tested at DSMZ on enterohaemolysin agar (PB5105A, OXOID, Wesel, Germany), strain DSM 30083T is enterohaemolysin-negative and thus does not belong to enterohemorrhagic serotype (enterohaemorrhagic E. coli, EHEC). The T phages T1-T7 did not lyse strain DSM 30083T cultivated on DSMZ medium 544 at 37°C.
Genome sequencing and annotation
Genome project history
The E. coli type strain genome was sequenced as part of the G enomic E ncyclopedia of B acteria and A rchaea (GEBA) project [4]. It was the only strain in the project that was chosen for genome sequencing due to its eminent prominence as a model organism and its value as a taxonomic reference strain and not selected according to the GEBA criteria for distinct phylogenetic location [4, 50]. Project information is found in the Genomes OnLine Database [1]. Draft sequencing, initial gap closure and annotation were performed by the DOE Joint Genome Institute (JGI) using state-of-the-art sequencing technology [51]. The Whole Genome Shotgun (WGS) sequence is deposited in Genbank and the Integrated Microbial Genomes database (IMG) [52]. A summary of the project information is shown in Table 2.
Growth conditions and DNA isolation
A culture of strain DSM 30083T was grown aerobically in DSMZ medium 1 [41] at 37°C. Genomic DNA was isolated using MasterPure Gram-Positive DNA Purification Kit (Epicentre MGP04100) following the standard protocol provided by the manufacturer but modified by incubation on ice over night on a shaker. DNA is available from DSMZ through the DNA Bank Network [53].
Genome sequencing and assembly
The genome was sequenced using a combination of 454-GS-FLX-Titanium and Illumina GAii platforms. Illumina contigs of a length greater than 800 bp were shredded into pieces of up to 1000 bp at 200 bp intervals prior to the velvet [54] assembly. An additional round of automated gap closure yielded a draft version of the genome sequence comprising 37 contigs. Further gap closure via primer walking and finishing with Consed [55] was conducted at LGC Genomics (Berlin) and resulted in three aligned contigs for the chromosome and one for the plasmid.
Genome annotation
Genes were identified using Prodigal [56] as part of the JGI genome annotation pipeline [57]. The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, UniProt, TIGR-Fam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Identification of RNA genes were carried out by using HMMER 3.0rc1 [58] (rRNAs) and tRNAscan-SE 1.23 [59] (tRNAs). Other non-coding genes were predicted using INFERNAL 1.0.2 [60]. Additional gene prediction analysis and functional annotation was performed within the Integrated Microbial Genomes - Expert Review (IMG-ER) platform [61] CRISPR elements were detected using CRT [62] and PILER-CR [63].
Genome properties
The genome statistics are provided in Table 3, Figure 3 and Figure 4. The genome of strain DSM 30083T has a total length of 5,038,133 bp and a G+C content of 50.6%. Of the 4,937 genes predicted, 4,762 were identified as protein-coding genes, and 175 as RNAs. The majority of the protein-coding genes were assigned a putative function (84.2%) while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Insights into the genome
Which E. coli genomes actually represent E. coli?
Since the focus of this study is the E. coli type strain DSM 30083T, we will only discuss genomic aspects related to this strain in the following. Indeed, only the availability of the type-strain genome enables one to assess with modern genome sequence-based taxonomic methods whether or not the large number of genome-sequenced E. coli strains actually belong to this species. The taxonomist’s main criterion for species affiliation is the 70% DNA:DNA hybridization (DDH) similarity threshold [64, 65], but here we use an improved modern variant of the method, which is based on intergenomic sequence distances [24, 25]. This approach retains consistency with the microbial species concept because the traditional DDH is, on average, closely mimicked, but digital DDH (dDDH) avoids the pitfalls of traditional DDH due to the much lower error rate in genome sequencing [26].
Figure 5 shows the dDDH similarities between DSM 30083T and a selection of 250 E. coli strains (see Additional file 2 for a full list) as well as outgroup genomes inferred using the Genome-to-Genome Distance Calculator [24], version 2 [25], which is based on the Genome BLAST Distance Phylogeny (GBDP) approach [24, 25]. Apparently, all strains identified as E. coli are within the 70% range of the type strain and hence need no reclassification (which would be the case for DDH values below the 70% threshold). The analysis also confirms that Shigella (within dDDH group IV) is placed within E. coli; this was already known from traditional DDH studies, yet the name Shigella was retained to not cause confusion in medical microbiology [39]. In accordance with the taxonomic classification, none of the strains from other Escherichia species yielded a dDDH similarity >70% (Figure 5).
Histogram of the digital DDH similarities between the type strain, DSM 30083T , and other genome-sequenced E. coli strains as well as outgroups. Interesting groups are marked by Roman numerals I-VII: Escherichia hermannii and Shimwellia blattae (I), E. fergusonii and E. albertii (II), E. sp. TW09308 (III), E. coli (IV-VII). Regarding the revised phylotypes from [66] (compare Figure 6), phylotype B2 is covered by dDDH groups V, VI, and VII with VII being the group containing (among other strains) type strain DSM 30083T itself and its closest relative E. coli S88. IV marks the biggest group which includes phylotypes A, B1, D1, D2, D3, E, F1, F2 and Shigella I, IIa, IIb and III. The full list of dDDH values and affiliation to phylotypes is contained in Additional file 2.
For easing the comparison with literature data, we used the phylotypes suggested by [67–69] and revised according to the sixth picture in [66], which reassigned strains to phylotypes in most cases where it was necessary to render them monophyletic in a phylogenetic analysis of E. coli core genes (based on nucleotide alignments of 1,278 core-genes from 186 E. coli genomes). We had to additionally split phylotype D into D1, D2 and D3 because this phylotype actually was distributed over three distinct clades in [66], and for analogous reasons had to split F into F1 and F2 and Shigella II into Shigella IIa and Shigella IIb. The affiliation of the genomes present in our data set to the original phylotypes, if available, and the revised ones is contained in Additional file 2. The affiliations of E. coli strains to serovars were collected from GOLD [1], those to pathovars from [1] and [70]; they are also listed in the supplement.
Regarding the dDDH groups V, VI and VII in Figure 5 containing the E. coli strains with a dDDH similarity to the type strain of around 85% or higher, those with an assigned revised phylotype uniformly belonged to phylotype B1. A histogram depicting the dDDH similarities between all strains used in this study is contained in Additional file 1.
Phylogenetic analysis with nucleotide GBDP
Figure 6 depicts a phylogenetic tree of the same strains inferred using GBDP, the highly reliable method [71] to calculate intergenomic distances, on which the inference of digital DDH values as shown in Figure 6 is also based [24, 25]. The branch support values in this tree (Figure 6) originate from pseudo-bootstrapping [25], a procedure which is known as conservative [72] and in the context of GBDP tends to underestimate branch support particularly for branches close to the tips [73]. Accordingly, the tree shows a well-supported backbone whereas terminal branches reveal less support.
Whole-genome phylogeny inferred using the latest GBDP version [25] and rooted with Escherichia albertii. Other outgroup organisms, separated by long branches, were removed to ease visualization (E. hermannii, Shimwellia blattae, and E. fergusonii) but are shown in Additional file 1. Numbers above branches are greedy-with-trimming pseudo-bootstrap [73] support values from 100 replicates if larger than 50%. Leaves are colored according to their affiliation to phylotypes. The outer circles show the affiliation of the strains to potential subspecies, pathovars and serovars (if the information was retrievable). Labels with numbers in square brackets are duplicates (due to label shortening) and refer to the following full strains/GenBank accessions: [1] CS6:LT+:ST+, [2] TW07815, [3] AM946981, [4] CP001509, [5] CP001665, [6] AFST00000000, [7] AFRH00000000, [8] K-12, MG1655 U00096, and [9] CM000960. An asterisk (*) indicates the K-12 wild type. ITOL [74] was used to visualize the tree inferred using FastME [75].
Nevertheless, the tree topology (Figure 6) shows all revised phylotypes as monophyletic, and some of them with high support. According to Figure 6 the type strain DSM 30083T is placed within phylotype B2 with E. coli S88 as its closest neighbor. The observation that the Shigella phylotypes occur in three different clades, but that these are all positioned within E. coli, together with earlier studies [76, 77] provides evidence against a recent study [78] which proposes Shigella spp. as a sister group of E. coli rather than at least one of its subgroups. A possible reason might be that [78] utilized an alignment-free genome signature (“CVTree”) approach which was recently shown to be less accurate than GBDP [71]. High (92%) support was achieved for a clade comprising phylotypes A, B1, C, Shigella I, Shigella IIa and Shigella IIb, and maximum support for a parent clade of that clade, also comprising phylotypes D2, D3, E and Shigella III. The serovars and pathovars, as far as attributable to the genomes used in this study, showed lower agreement with the tree topology. This might be due to the highly diverse adaptive paths present in E. coli [77].
Phylogenetic analysis of proteome sequences
The genome sequences of a subset of 50 representative genome-sequenced strains were phylogenetically investigated in a complementary analysis using the DSMZ phylogenomics pipeline as previously described [79–86] using NCBI BLAST [87], OrthoMCL [88], MUSCLE [89], RASCAL [90], GBLOCKS [91] and MARE [92] to generate concatenated alignments of distinct selections of genes (supermatrices). Maximum likelihood (ML) [93] and maximum parsimony (MP) [94, 95] trees were inferred from the data matrices with RAxML [96, 97] and PAUP* [98], respectively, as previously described [79–86].
The topology of the ML MARE-filtered supermatrix analysis is shown in Figure 7 together with ML and MP bootstrap support values from all supermatrix analyses if larger than 60%. Support was maximum (100%) for the majority of branches under ML and MP (Figure 7). Again, in this tree all phylotypes are represented as monophyla with the sole exception of B1, which was revealed only in the core-gene analysis, much like in [66]. A further difference to the 2012 study [66] and the GBDP tree (Figure 6) is that Shigella phylotypes I, IIa, IIb and III formed a clade together; again this clade was not visible in the core-gene tree. In our view, trying distinct ways to generate supermatrices has the strong advantage that branches that are sensitive to gene selection can be revealed [79–86]. Whereas the above-mentioned groups are instable in this respect, others such as the group comprising phylotypes A, B1, E and all Shigella strains yield maximum support under all assessed gene selections; this large clade also obtained 100% support with GBDP (Figure 6). Average branch support under ML and MP, respectively, was 91.72/87.62% using the core genes only (101,755 variable, 21,474 parsimony-informative characters), 94.04/97.64% using the MARE-filtered supermatrix (285,814/99,071) and 90.3/97.49% using the entire supermatrix (456,246/153,146). This is largely in agreement with the tendency observed in previous studies using the same phylogenomics pipeline that more characters simply yield higher support, despite the frequent concerns regarding horizontal gene transfer [99], but might also indicate advantages of the removal of phylogenetically uninformative genes with MARE [92].
Phylogenetic tree inferred from the MARE-filtered supermatrix under the maximum likelihood (ML) criterion and rooted with E. hermannii NBRC 105704. Branch lengths within the outgroup were shortened to improve visualization. The branches are scaled in terms of the expected number of substitutions per site. Numbers above / below the branches (from left to right) are bootstrapping support values (if larger than 60%) from (i) ML MARE-filtered supermatrix; (ii) maximum-parsimony (MP) MARE-filtered supermatrix; (iii) ML “full” supermatrix, (iv) MP “full” supermatrix, (v) ML core-genes; (vi) MP core-genes analysis. Dots indicate branches with maximum support under all settings. Numbers in square brackets refer to further strain information as listed in the caption of Figure 6.
Phylogenetic analysis of gene and ortholog content
The clusters of orthologs as inferred with OrthoMCL, as well as clusters of homologs inferred using re-implementation of the TribeMCL [100] algorithm as previously described [79–86], were converted to presence-absence matrices for phylogenetic inference using ML and MP. The topology of the MP ortholog-content analysis is shown in Figure 8 together with MP and ML bootstrap support values from ortholog-content and gene-content analyses if larger than 60%. In contrast to the GBDP (Figure 6) and supermatrix (Figure 7) analyses, E. coli forms a sister group of Shigella spp., but with at most moderate (80%) support. Similarly, the clade containing both is at most moderately supported. Support for a monophyletic Shigella is high, however (98-100%). The phylotypes are revealed as monophyletic except for F1 and B1 (with strong support against them forming a clade, respectively).
Phylogeny inferred from the ortholog-content matrix under the maximum parsimony (MP) criterion and rooted with E. hermannii NBRC 105704. The branches are scaled in terms of the minimum number of substitutions (DELTRAN optimization). Numbers above/below the branches (from left to right) are bootstrapping support values (if larger than 60%) from (i) MP ortholog-content matrix; (ii) maximum-likelihood (ML) ortholog-content matrix; (iii) MP gene-content matrix; (iv) ML gene-content matrix analysis. Dots indicate branches with maximum support under all settings.
The within-species difference of genomic G+C content
The G+C content of 50.6% inferred from the genome sequence is in agreement with the value of 50.7 ± 0.6 mol% determined for strain DSM 30083T by Albuquerque et al. [101], but differs slightly from the G+C content of 51.0-51.7 mol%, determined from deposit ATCC 11775T [29]. The G+C content range of E. coli strains was reported as 48.5-52.1 mol% [29], in conflict with more recent results [26]. Thus affiliation to E. coli was also assessed by calculating the genomic G+C content of all 251 strains in the data set and the difference to the G+C content of the type strain, DSM 30083T. Results shown in Figure 9 are in agreement with the result from [26] that within-species differences in the G+C content are almost exclusively below 1%. As expected, E. coli cannot be distinguished from the other Escherichia species based on G+C content.
Histogram of the differences in genomic G+C content between the E. coli type strain and the other 250 strains contained in the data set. In accordance to a within-species difference of at most 1% in the G+C content [26], none of the differences between the distinct strains of E. coli are above that threshold. The G+C differences to E. hermannii NBRC 105704 (3.4%, “A”) and Shimwellia blattae DSM 4481 (5.9%, “B”) are considerably larger, whereas E. albertii, E. fergusonii and “Escherichia sp. TW09308” (which are also phylogenetically more close to E. coli; see Figure 6 and Figure 7) cannot be distinguished from E. coli using the G+C content.
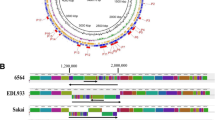
The 131-kb plasmid of E. coli DSM 30083T
The E. coli type strain DSM 30083T contains a single circular incFII-type plasmid with a size of 131,289 bp and a G+C content of 49.3% (Figure 4). A homologous plasmid that just exhibits an inversion of 15 kb and an indel (insertion/deletion) of 3 kb is present in the closest relative E. coli strain S88 (CU928146). The 131-kb plasmid harbors a type IV secretion system and a highly syntenous conjugative plasmid has been identified in a multidrug-resistent Salmonella enterica strain CVM29188 (NC_011076) [102] thus providing strong evidence of natural interspecies exchange of the extrachromosomal element.
Physiological discrimination of E. coli DSM 30083Tand DSM 18039
Since the genomes of both E. coli strains DSM 30083T and K-12 MG1655 (=DSM 18039) fall into strongly separated clusters, the question of phenotypic differences between the type strain and the widely used laboratory strain arises, too. We thus also investigated the substrate spectrum of using PM-01 and PM-02 microplates as described above (see also Additional file 1). In contrast to DSM 30083T, DSM 18039 was positive for dulcitol, D-xylose, α-keto-glutaric acid, m- tartaric acid, α-hydroxy-butyric acid, 5-keto-d-gluconic acid, but negative for l-glutamic acid, d-glucosaminic acid, tween 20, tween 40, mono-methyl succinate, N-acetyl-d-galactosamine, d-arabinose, d-raffinose, l-sorbose, d-tagatose. On API 20E strips (see Additional file 1) strain DSM 18039T in contrast to E. coli 30083T was negative for l-ornithine. A unique diagnostic trait of all completely sequenced K-12 strains that allow the discrimination from other E. coli isolates is a deletion of 3,205 bp in the aga gene cluster that is required for the conversion of N-acetyl-d-galactosamine [103].
Subdivision of E. coli revisited
As shown above, after a small number of revisions as conducted in [66] and partially in this study, the proposed phylotypes of E. coli appear monophyletic in the phylogenetic analyses of genome-scale data. The sole exception is phylotype B1, whose monophyly is confirmed in Figure 6 but shows a sensitivity to gene selection in analyses of proteome sequences (Figure 7). The additional question arises, however, whether or not the phylotypes are not only monophyletic but also are comparable to each other with respect to the level of character divergence within each group. This would be advantageous for (formal or informal) classification, as can easily be shown by a comparison with the 70% DDH rule for delineating bacterial species. There is, unfortunately, no guarantee that the set of strains in the 70% (d)DDH range of a type strain form a monophyletic group unless the distances are ultrametric [26]. But on the other hand, in contrast to the monophyly criterion itself, the consequent application of the 70% DDH rule by construction yields groups with a similar upper bound of character divergence. The same reasoning also holds for organisms not covered by the Bacteriological Code. For instance, whereas birds, mammals and primates are all monophyletic according to current knowledge, comparing birds and mammals regarding, say, species numbers makes much more sense than comparing birds and primates.
To assess the homogeneity of the revised E. coli phylotypes, some of their cluster statistics were calculated with OPTSIL [104] version 1.5 and the matrix of intergenomic distances used for inferring dDDH values (Figure 5). Average within-cluster distances ranged between 0.00098 and 0.01571 with a median of 0.00503, whereas maximum within-cluster distances ranged between 0.00121 and 0.02199 with a median of 0.01444. Further, clustering optimization as implemented in OPTSIL was conducted using the revised phylotypes as reference partition; details are found in Additional file 3. The maximum agreement with the reference partition was obtained for a combination of clustering parameters that yielded 32 clusters, way more than the number of phylotypes plus outgroups that were input into clustering optimization.
This analysis shows that the phylotypes of E. coli, even if revised to obtain monophyly of all phylotypes in the phylogenetic analyses of genome-scale data as conducted in [66] and this study (Figure 6), are not homogeneous regarding their divergence as measured using genome-scale nucleotide data. This can also be shown indirectly by comparing the phylotypes to a clustering conducted with the slightly higher distance threshold of 0.0242, which corresponds to 79.3% dDDH. The tree in Figure 6 is annotated with this clustering, too; it yields five clusters, four of which obtain GBDP pseudo-bootstrap values between 98% and 100%. Four of these clusters directly correspond to one phylotype, respectively, namely B2, D1, F1 and F2, whereas the fifth cluster comprises all remaining phylotypes, including all Shigella spp. (Figure 6). Interestingly, in contrast to some phylotypes, this cluster is supported in proteome-based trees under all investigated settings (Figure 7). It is not supported by the gene-content based phylogenies (Figure 8), but these neither yield support against this cluster. Thus if measured from genome-scale nucleotide data the phylotypes B2, D1, F1 and F2, as well as the combination of all remaining clusters have approximately the same level of divergence, respectively.
Delineation of subspecies revisited
Bacterial subspecies were traditionally not determined based on a distance or similarity threshold, but on a qualitative assessment of few selected phenotypic characters [65, 105, 106]. A quotation from [64] is worth reproducing here: “Subspecies designations can be used for genetically close organisms that diverge in phenotype. There is some evidence, based on frequency distribution of ΔT m values in DNA hybridization, that the subspecies concept is phylogenetically valid. (…) There is a need for further guidelines for designation of subspecies.” Particularly because the availability of complete genome sequences allows for the transition to genome-based taxonomy, yielding to a considerable increase in phylogenetic resolution [99], rules for a genome-based, quantitative approach to subspecies delineation in analogy to the 70% (d)DDH threshold for the delineation of species [24, 25, 65], would be desirable.
However, as emphasized in [26], inconsistencies can occur when distance or similarity thresholds are used and the underlying distances specifically deviate from ultrametricity. These potential pitfalls are a general consequence of the direct use of pairwise distances or similarities (which is not a phylogenetic method) for assessing taxonomic affiliations [104] and not directly related to traditional or digital DDH. Fewer taxonomic problems are expected when comparisons between two non-type strains are avoided (which is necessary for reasons of nomenclature anyway), but this does not entirely prevent pitfalls [26]. Nevertheless, whether paradoxes really occur in practice depends on the distance threshold and the specific deviation of the data under study from the ultrametric condition [26]. Hence, if a threshold for delineating bacterial subspecies is of interest, it makes sense to choose it so as to minimize the potential of taxonomic inconsistencies related to non-ultrametric data as far as possible. This can be done for bacterial subspecies precisely because by tradition they have not been determined based on a distance or similarity threshold, in contrast to the species rank, hence such a threshold can now be carefully chosen based on the above-mentioned principles.
Using the E. coli data as starting point, augmented by the data set used in [26] containing completely sequenced genomes for 105 genera of Archaea and Bacteria, in addition to criteria from the literature we have devised a criterion called “clustering consistency” for optimizing thresholds for sub-specific bacterial lineages. Compared to the analysis of frequency distributions of (d)DDH values as mentioned in [99], this approach has the advantage that it directly addresses how to best cluster the sequences. The analyses described in detail in Additional file 3 show that regarding within-species clustering consistency a distance threshold corresponding to 79-80% dDDH makes most sense for both the E. coli and the 105-genera Archaea and Bacteria data sets. In addition to clustering consistency, a value around 80% has a couple of other advantages. For instance, it is sufficiently larger than the species boundary at 70% but nevertheless does not yield too many subspecies if applied strictly. This is particularly important regarding the low number of currently described subspecies in the literature, which in our view makes it also impossible to estimate dDDH subspecies boundaries from the currently validly named subspecies. Furthermore, values between 90% and 95% dDDH could be reserved in the future for taxonomic ranks such as “variety”. Finally, values approaching 100% are unsuitable because they might represent distinct clones or deposits of the same strain or even genome sequences obtained several times from the same strain.
Taxonomic consequences for E. coli?
As mentioned above, E. coli is an attractive example for the application of the 79-80% dDDH rule (Figure 6). Hence, the description of subspecies of E. coli is the next logical consequence. Regarding practice, it is noteworthy that the already established detection of phylotypes [67–69] will help detecting the subspecies, too, because the (revised) phylotypes are either identical to subspecies or to subsets of subspecies (Figure 6). Furthermore, even incompletely sequenced genomes can be used to detect the subspecies by the comparison with the type strains using the GGDC server [24, 25]. Apparently, Shigella spp. would not only be placed within E. coli [107] but even embedded within one of the subspecies defined at the 79-80% dDDH boundary (Figure 6). Crucially, this changes nothing regarding the status of Shigella: if this name is to be retained not to cause confusion in medical microbiology anyway [39], it simply does not matter whether or not it otherwise would be placed entirely within E. coli or even entirely within a yet to be established subspecies of E. coli.
However, the placement of Shigella yields yet another problem for the division of E. coli into subspecies. An approach to describe subspecies for E. coli could start with the largest cluster in Figure 6, which contains most of the genome-sequenced strains including strain K-12, but also all strains of Shigella. Following the guidelines of the Bacteriological Code (1990 revision) [2] the type strain of this subspecies would be strain NewcastleT (=NCTC 4837T) representing E. coli subsp. dysenteriae (Shiga 1897) Castellani and Chalmers 1919, with strain U5/41T automatically becoming the type strain of E. coli subsp. coli (Shiga 1897) Castellani and Chalmers 1919. Thus establishing this subspecies of E. coli would taxonomically conflict with the purpose of retaining Shigella [39], hence we refrain from proposing taxonomic consequences here. The dDDH boundary suggested in this study for delineating subspecies might nevertheless be of use on many other groups of Bacteria and Archaea that are not hampered by similar (taxonomic) constraints.
Conclusions
This study presents the genome sequence for the E. coli type strain DSM 30083T, whose marked physiological and genomic differences from the model bacterium E. coli K-12 are reviewed in detail. A phylogenomic analysis of 250 E. coli strains reveals that their arrangement into the phylotypes suggested in the literature, even though they mostly appear monophyletic, does not yield a uniform level of character divergence. We thus propose an alternative arrangement and discuss it in the context of the subspecies rank. This is of special interest because bacterial subspecies were traditionally not determined based on a distance or similarity threshold but an approach to quantitatively delineate them has been requested in the literature. Based on an investigation of genome-sequenced strains from > 100 genera, including E. coli, and the criterion of clustering consistency, we suggest a boundary of 79-80% dDDH for delineating subspecies within Bacteria and Archaea. Such dDDH-based subspecies delineation is available via the GGDC web service.
In E. coli, the criterion yields five subspecies, one of which includes strain 30083T and is identical to phylogroup B2. Strain K-12, together with Shigella and the majority of E. coli strains, belongs to another subspecies. Issues of nomenclature prevent taxonomic consequences in E. coli, but the methodology applied here is of general interest for bacterial subspecies delineation.
References
Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC: The Genomes OnLine Database (GOLD) v. 4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res 2012, 40: D571–9. 10.1093/nar/gkr1100
Lapage SP, Sneath PHA, Lessel EF, Skerman VBD, Seeliger HPR, Clark WA: International Code of Nomenclature of Bacteria, 1990 Revision. Washington DC: ASM Press; 1992.
Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis N, Kirkpatrick H, Goeden M, Rose D, Mau B, Shao Y: The complete genome sequence of Escherichia coli K-12. Science 1997, 277: 1453–65. 10.1126/science.277.5331.1453
Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, Hooper SD, Pati A, Lykidis A, Spring S, Anderson IJ, D'haeseleer P, Zemla A, Singer M, Lapidus A, Nolan M, Copeland A, Han C, Chen F, Cheng J-F, Lucas S, Kerfeld C, Lang E, Gronow S, Chain P, Bruce D, Rubin EM, et al.: A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea . Nature 2009, 462: 1056–60. 10.1038/nature08656
Escherich T: Die Darmbakterien des Säuglings und ihre Beziehungen zur Physiologie der Verdauung. Stuttgart: Ferdinand Enke; 1886:63–74.
Judicial Commission of the International Committee on Bacterial Nomenclature: Conservation of the family name Enterobacteriaceae , of the name of the type genus, and designation of the type species. Int Bull Bacteriol Nomencl Taxon 1958, 8: 73–4.
Skerman V, McGowan V, Sneath P: Approved lists of bacterial names. Int J Syst Bacteriol 1980, 30: 225–420. 10.1099/00207713-30-1-225
Kauffmann F: Zur Serologie der Coli-Gruppe. Acta Pathol Microbiol Scand 1944, 21: 20–45.
Editorial Board (for the Judicial Commission of the International Committee on Bacteriological Nomenclature): Opinion 26: designation of neotype strains (cultures) of type species of the bacterial genera Salmonella, Shigella, Arizona, Escherichia, Citrobacter and Proteus of the family Enterobacteriaceae . Int J Syst Evol Microbiol 1963, 13: 35–6.
Ørskov F, Ørskov I: 2. Serotyping of Escherichia coli . In Methods in Microbiology. Volume 14. Edited by: Bergan T. London: Academic Press; 1984:43–112.
Filannino P, Azzi L, Cavoski I, Vincentini O, Rizzello CG, Gobbetti M, Di Cagno R: Exploitation of the health-promoting and sensory properties of organic pomegranate (Punica granatum L.) juice through lactic acid fermentation. Int J Food Microbiol 2013, 163: 184–92. 10.1016/j.ijfoodmicro.2013.03.002
Schumann P, Pukall R: The discriminatory power of ribotyping as automatable technique for differentiation of bacteria. Syst Appl Microbiol 2013, 36: 369–75. 10.1016/j.syapm.2013.05.003
Farnleitner A, Kreuzinger N, Kavka G, Grillenberger S, Rath J, Mach R: Simultaneous detection and differentiation of Escherichia coli populations from environmental freshwaters by means of sequence variations in a fragment of the β- D -glucuronidase gene . Appl Environ Microbiol 2000, 66: 1340–6. 10.1128/AEM.66.4.1340-1346.2000
Tee TW, Chowdhury A, Maranas CD, Shanks JV: Systems metabolic engineering design: Fatty acid production as an emerging case study. Biotechnol Bioeng 2014, 111: 849–57. 10.1002/bit.25205
Wen M, Bond-Watts BB, Chang MCY: Production of advanced biofuels in engineered. E. coli . Curr Opin Chem Biol 2013, 17: 472–9. 10.1016/j.cbpa.2013.03.034
Rosano GL, Ceccarelli EA: Recombinant protein expression in Escherichia coli : advances and challenges . Front Microbiol 2014, 5: 172.
Donovan C, Bramkamp M: Cell division in Corynebacterineae . Front Microbiol 2014, 5: 132.
Kuzminov A: The chromosome cycle of prokaryotes. Mol Microbiol 2013, 90: 214–27.
Kang Z, Zhang C, Zhang J, Jin P, Zhang J, Du G, Chen J: Small RNA regulators in bacteria: powerful tools for metabolic engineering and synthetic biology. Appl Microbiol Biotechnol 2014, 98: 3413–24. 10.1007/s00253-014-5569-y
Whitfield C, Roberts I: Structure, assembly and regulation of expression of capsules in Escherichia coli . Mol Microbiol 1999, 31: 1307–19. 10.1046/j.1365-2958.1999.01276.x
Cooper K, Mandrell R, Louie J, Korlach J, Clark T, Parker C, Huynh S, Chain P, Ahmed S, Carter M: Comparative genomics of enterohemorrhagic Escherichia coli O145:H28 demonstrates a common evolutionary lineage with Escherichia coli O157:H7 . BMC Genomics 2014, 15: 17. 10.1186/1471-2164-15-17
Allocati N, Masulli M, Alexeyev MF, Di Ilio C: Escherichia coli in Europe: an overview . Int J Environ Res Public Health 2013, 10: 6235–54. 10.3390/ijerph10126235
Kaper JB, Nataro JP, Mobley HL: Pathogenic Escherichia coli . Nat Rev Microbiol 2004, 2: 123–40. 10.1038/nrmicro818
Auch AF, Von Jan M, Klenk HP, Göker M: Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2010, 2: 117–34. 10.4056/sigs.531120
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M: Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 2013, 14: 60. 10.1186/1471-2105-14-60
Meier-Kolthoff JP, Klenk HP, Göker M: Taxonomic use of the G+C content and DNA:DNA hybridization in the genomic age. Int J Syst Evol Microbiol 2014, 64: 352–6. 10.1099/ijs.0.056994-0
Göker M, Cleland D, Saunders E, Lapidus A, Nolan M, Lucas S, Hammon N, Deshpande S, Cheng J-F, Tapia R, Han C, Goodwin L, Pitluck S, Liolios K, Pagani I, Ivanova N, Mavromatis K, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang Y-J, Jeffries C, Detter J, Beck B, Woyke T, Bristow J, Eisen J, Markowitz V, et al.: Complete genome sequence of Isosphaera pallida type strain (IS1BT ) . Stand Genomic Sci 2011, 4: 63–71. 10.4056/sigs.1533840
Welch RA: 3.3.3 The Genus Escherichia . In The Prokaryotes. Third edition, Volume 6. Edited by: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. Berlin: Springer; 2005:62–71.
Scheutz F, Strockbine NA: Genus I. Escherichia Castellani and Chalmers 1919 . In Bergey’s Manual of Systematic Bacteriology. Second edition, Volume 2 (The Proteobacteria). Edited by: Brenner DJ, Krieg NR, Staley JT. New York: Springer; 2005:607–24.
Koser SA: Utilization of the salts of organic acids by the colon-aerogenes group. J Bacteriol 1923, 8: 493–520.
Topley WWC, Wilson GS: The Principles of Bacteriology and Immunity. 2nd edition. 1936.
Huys G, Cnockaert M, Janda JM, Swings J: Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children . Int J Syst Evol Microbiol 2003, 53: 807–10. 10.1099/ijs.0.02475-0
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, De Vos P, de Pamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glockner FO, Goldstein P, Guralnick R, Haft D, Hancock D, et al.: The minimum information about a genome sequence (MIGS) specification. Nature Biotechnol 2008, 26: 541–7. 10.1038/nbt1360
Field D, Amaral-Zettler L, Cochrane G, Cole JR, Dawyndt P, Garrity GM, Gilbert J, Glöckner FO, Hirschman L, Karsch-Mizrachi I, Klenk H-P, Knight R, Kottmann R, Kyrpides N, Meyer F, San Gil I, Sansone S-A, Schriml LM, Sterk P, Tatusova T, Ussery DW, White O, Wooley J: The Genomic Standards Consortium. PLoS Biol 2011, 9: e1001088. 10.1371/journal.pbio.1001088
Woese CR, Kandler O, Weelis ML: Towards a natural system of organisms. Proposal for the domains Archaea and Bacteria . Proc Natl Acad Sci U S A 1990, 87: 4576–9. 10.1073/pnas.87.12.4576
Garrity GM, Bell JA, Lilburn T: Phylum XIV. Proteobacteria phyl nov. In Bergey’s Manual of Systematic Bacteriology. Second edition, Volume 2 (The Proteobacteria part B The Gammaproteobacteria). Edited by: Brenner DJ, Krieg NR, Stanley JT, Garrity GM. New York: Springer; 2005:1.
Garrity GM, Bell JA, Lilburn T: Class III. Gammaproteobacteria class. nov. In Bergey’s Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B. Edited by: Garrity GM, Brenner DJ, Krieg NR, Staley JT. New York: Springer; 2005:1.
Williams KP, Kelly DP: Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int J Syst Evol Microbiol 2013, 63: 2901–2906. doi:10.1099/ijs.0.049270–0 10.1099/ijs.0.049270-0
Brenner DJ: Family I. Enterobacteriaceae Rahn 1937, Nom. fam. cons. Opin. 15, Jud. Com. 1958, 73; Ewing, Farmer, and Brenner 1980, 674; Judicial Commission 1981, 104. In Bergey’s Manual of Systematic Bacteriology. First edition, Volume 1. Edited by: Krieg NR, Holt JG. Baltimore: The Williams & Wilkins Co; 1984:408–20.
Castellani A, Chalmers AJ: Manual of Tropical Medicine Third edition. New York: Williams Wood and Co; 1919:941–2.
List of growth media used at the DSMZ http://www.dsmz.de/
BAuA: TRBA 466: Classification of Bacteria and Archaea in Risk Groups. Berlin: BAuA; 2010:93.
Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry J, Davis A, Dolinski K, Dwight S, Eppig J, Harris M, Hill D, Issel-Tarver L, Kasarskis A, Lewis S, Matese J, Richardson J, Ringwald M, Rubin G, Sherlock G, Consortium GO: Gene ontology: tool for the unification of biology. Nat Genet 2000, 25: 25–9. 10.1038/75556
Vaas LAI, Sikorski J, Michael V, Göker M, Klenk HP: Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS ONE 2012, 7: e34846. 10.1371/journal.pone.0034846
Vaas LAI, Sikorski J, Hofner B, Fiebig A, Buddruhs N, Klenk HP, Göker M: opm: an R package for analysing OmniLog phenotype microarray data. Bioinformatics 2013, 29: 1823–4. 10.1093/bioinformatics/btt291
R Development Core Team R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014.
Chang YF, Feingold DS: D-glucaric acid and galactaric acid catabolism by Agrobacterium tumefaciens . J Bacteriol 1970, 102: 85–96.
Boer H, Maaheimo H, Koivula A, Penttila M, Richard P: Identification in Agrobacterium tumefaciens of the D -galacturonic acid dehydrogenase gene . Appl Microbiol Biotechnol 2010, 86: 901–9. 10.1007/s00253-009-2333-9
Xiao Z, Xu P: Acetoin metabolism in bacteria. Crit Rev Microbiol 2007, 33: 127–40. 10.1080/10408410701364604
Göker M, Klenk HP: Phylogeny-driven target selection for large-scale genome-sequencing (and other) projects. Stand Genomic Sci 2013, 8: 360–74. 10.4056/sigs.3446951
Mavromatis K, Land ML, Brettin TS, Quest DJ, Copeland A, Clum A, Goodwin L, Woyke T, Lapidus A, Klenk HP, Cottingham RW, Kyrpides NC: The fast changing landscape of sequencing technologies and their impact on microbial assemblies and annotations. PLoS ONE 2012, 7: e48837. 10.1371/journal.pone.0048837
Markowitz VM, Chen I-M A, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC: IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 2012, 40: D115–22. 10.1093/nar/gkr1044
Gemeinholzer B, Dröge G, Zetzsche H, Haszprunar G, Klenk HP, Güntsch A, Berendsohn WG, Wägele JW: The DNA Bank Network: the start from a German initiative. Biopreserv Biobank 2011, 9: 51–5. 10.1089/bio.2010.0029
Zerbino DR, Birney E: Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008, 18: 821–9. 10.1101/gr.074492.107
Gordon D, Abajian C, Green P: Consed: a graphical tool for sequence finishing. Genome Res 1998, 8: 195–202. 10.1101/gr.8.3.195
Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ: Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010, 11: 119. 10.1186/1471-2105-11-119
Mavromatis K, Ivanova NN, Chen IM, Szeto E, Markowitz VM, Kyrpides NC: The DOE-JGI standard operating procedure for the annotations of microbial genomes. Stand Genomic Sci 2009, 1: 63–7. 10.4056/sigs.632
Finn DR, Clements J, Eddy SR: HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 2011, 39: W29–37. 10.1093/nar/gkr367
Lowe TM, Eddy SR: tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997, 25: 955–64. 10.1093/nar/25.5.0955
Nawrocki EP, Kolbe DL, Eddy SR: Infernal 1.0: inference of RNA alignments. Bioinformatics 2009, 25: 1335–7. 10.1093/bioinformatics/btp157
Markowitz VM, Ivanova NN, Chen IMA, Chu K, Kyrpides NC: IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 2009, 25: 2271–8. 10.1093/bioinformatics/btp393
Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P: CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 2007, 8: 209. 10.1186/1471-2105-8-209
Edgar RC: PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformatics 2007, 8: 18. 10.1186/1471-2105-8-18
Wayne L, Brenner D, Colwell R, Grimont P, Kandler O, Krichevsky M, Moore L, Moore W, Murray R, Stackebrandt E, Starr M, Truper H: Report of the Ad Hoc Committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 1987, 37: 463–4. 10.1099/00207713-37-4-463
Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P: Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 2010, 60: 249–66. 10.1099/ijs.0.016949-0
Kaas RS, Friis C, Ussery DW, Aarestrup FM: Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics 2012, 13: 577. 10.1186/1471-2164-13-577
Clermont O, Bonacorsi S, Bingen E: Rapid and simple determination of the Escherichia coli phylogenetic group . Appl Environ Microbiol 2000, 66: 4555–8. 10.1128/AEM.66.10.4555-4558.2000
Clermont O, Gordon DM, Brisse S, Walk ST, Denamur E: Characterization of the cryptic Escherichia lineages: rapid identification and prevalence. Environ Microbiol 2011, 13: 2468–77. 10.1111/j.1462-2920.2011.02519.x
Clermont O, Christenson JK, Denamur E, Gordon DM: The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups . Environ Microbiol Rep 2013, 5: 58–65. 10.1111/1758-2229.12019
Sahl JW, Morris CR, Rasko DA: Comparative genomics of pathogenic Escherichia coli . In Escherichia coli: Pathotypes and Principles of Pathogenesis. Second edition. Edited by: Donnenberg MS. London: Academic Press; 2013.
Patil KR, McHardy AC: Alignment-free genome tree inference by learning group-specific distance metrics. Genome Biol Evol 2013, 5: 1470–84. 10.1093/gbe/evt105
Thorne JLL, Kishino H: Freeing phylogenies from artifacts of alignment. Mol Biol Evol 1992, 9: 1148–62.
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M: Highly parallelized inference of large genome-based phylogenies. Concurrency Comput Pract Ex 2014. in press
Letunic I, Bork P: Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 2011, 39: W475–8. 10.1093/nar/gkr201
Desper R, Gascuel O: Fast and accurate phylogeny minimum-evolution principle. J Comput Biol 2002, 9: 687–705. 10.1089/106652702761034136
Lukjancenko O, Wassenaar TM, Ussery DW: Comparison of 61 sequenced Escherichia coli genomes . Microb Ecol 2010, 60: 708–20. 10.1007/s00248-010-9717-3
Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, El Karoui M, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguenec C, Lescat M, Mangenot S, Martinez-Jehanne V, Matic I, Nassif X, Oztas S, et al.: Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genetics 2009, 5: e1000344. 10.1371/journal.pgen.1000344
Zuo G, Xu Z, Hao B: Shigella strains are not clones of Escherichia coli but sister species in the genus Escherichia . Genomics Proteomics Bioinformatics 2013, 11: 61–5. 10.1016/j.gpb.2012.11.002
Abt B, Han C, Scheuner C, Lu M, Lapidus A, Nolan M, Lucas S, Hammon N, Deshpande S, Cheng J-F, Tapia R, Goodwin L, Pitluck S, Mavromatis K, Mikhailova N, Huntemann M, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Brambilla E, Rohde M, Spring S, Gronow S, Göker M, Woyke T, Bristow J, Eisen J, Markowitz V, et al.: Complete genome sequence of the termite hindgut bacterium Spirochaeta coccoides type strain (SPN1T ), reclassification in the genus Sphaerochaeta as Sphaerochaeta coccoides comb. nov. and emendations of the family Spirochaetaceae and the genus Sphaerochaeta . Stand Genomic Sci 2012, 6: 194–209. 10.4056/sigs.2796069
Abt B, Göker M, Scheuner C, Han C, Lu M, Misra M, Lapidus A, Nolan M, Lucas S, Hammon N, Deshpande S, Chang J-F, Tapia R, Goodwin L, Pitluck S, Liolios K, Pagani I, Ivanova N, Mavromatis K, Mikhailova N, Huntemann M, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Brambilla E-M, Rohde M, Spring S, Gronow S, et al.: Genome sequence of the thermophilic fresh-water bacterium Spirochaeta caldaria type strain (H1T ), reclassification of Spirochaeta caldaria and Spirochaeta stenostrepta , and Spirochaeta zuelzerae in the genus Treponema as Treponema caldaria comb. nov., Treponema stenostrepta comb. nov., and Treponema zuelzerae comb. nov., and emendation of the genus Treponema . Stand Genomic Sci 2013, 8: 88–105. 10.4056/sigs.3096473
Anderson I, Scheuner C, Göker M, Mavromatis K, Hooper SD, Porat I, Klenk H-P, Ivanova N, Kyrpides N: Novel insights into the diversity of catabolic metabolism from ten haloarchaeal genomes. PLoS ONE 2011, 6: e20237. 10.1371/journal.pone.0020237
Frank O, Pradella S, Rohde M, Scheuner C, Klenk H-P, Göker M, Petersen J: Complete genome sequence of the Phaeobacter gallaeciensis type strain CIP 105210T (= DSM 26640T = BS107T). Stand Genomic Sci 2014., 9: in press
Göker M, Scheuner C, Klenk HP, Stielow JB, Menzel W: Codivergence of mycoviruses with their hosts. PLoS ONE 2011, 6: e22252. 10.1371/journal.pone.0022252
Spring S, Scheuner C, Lapidus A, Lucas S, Del Rio TG, Tice H, Copeland A, Cheng J-F, Chen F, Nolan M, Saunders E, Pitluck S, Liolios K, Ivanova N, Mavromatis K, Lykidis A, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang Y-J, Jeffries CD, Goodwin L, Detter JC, Brettin T, Rohde M, Göker M, Woyke T, Bristow J, et al.: The genome sequence of Methanohalophilus mahii SLPT reveals differences in the energy metabolism among members of the Methanosarcinaceae inhabiting freshwater and saline environments . Archaea 2010, 2010: 690737.
Stackebrandt E, Scheuner C, Göker M, Schumann P: Family Intrasporangiaceae. In The Prokaryotes – Actinobacteria. Fourth edition. Edited by: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F. Berlin: Springer; 2014. in press
Verbarg S, Göker M, Scheuner S, Schumann P, Stackebrandt E: The families Erysipelotrichaceae emend., Coprobacillaceae fam. nov., and Turicibacteraceae fam. nov. In The Prokaryotes. Fourth edition. Edited by: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F. Berlin: Springer; 2014. in press
Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997, 25: 3389–402. 10.1093/nar/25.17.3389
Li L, Stoeckert CJ Jr, Roos DS: OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 2003, 13: 2178–89. 10.1101/gr.1224503
Edgar RC: MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004, 32: 1792–7. 10.1093/nar/gkh340
Thompson JD, Thierry J-CC, Poch O: RASCAL: rapid scanning and correction of multiple sequence alignments. Bioinformatics 2003, 19: 1155–61. 10.1093/bioinformatics/btg133
Castresana J: Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000, 17: 540–52. 10.1093/oxfordjournals.molbev.a026334
Meusemann K, von Reumont BM, Simon S, Roeding F, Strauss S, Kuck P, Ebersberger I, Walzl M, Pass G, Breuers S, Achter V, von Haeseler A, Burmester T, Hadrys H, Wagele JW, Misof B: A phylogenomic approach to resolve the arthropod tree of life. Mol Biol Evol 2010, 27: 2451–64. 10.1093/molbev/msq130
Felsenstein J: Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 1981, 17: 368–76. 10.1007/BF01734359
Fitch WM: Toward defining the course of evolution: minimum change on a specified tree topology. Syst Zool 1977, 20: 406–16.
Goloboff PA: Parsimony, likelihood, and simplicity. Cladistics 2003, 19: 91–103. 10.1111/j.1096-0031.2003.tb00297.x
Stamatakis A: RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22: 2688–90. 10.1093/bioinformatics/btl446
Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A: How many bootstrap replicates are necessary? J Comput Biol 2010, 17: 337–54. 10.1089/cmb.2009.0179
Swofford DL: PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0 b10. Sunderland, MA: Sinauer & Associates; 2002.
Klenk HP, Göker M: En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 2010, 33: 175–82. 10.1016/j.syapm.2010.03.003
Enright AJ, van Dongen SM, Ouzounis CA: An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 2002, 30: 1575–84. 10.1093/nar/30.7.1575
Albuquerque L, Rainey FA, Fernanda Nobre M, da Costa MS: Hydrotalea sandarakina sp. nov., isolated from a hot spring runoff, and emended descriptions of the genus Hydrotalea and the species Hydrotalea flava. Int J Syst Evol Microbiol 2012, 62: 1603–8. 10.1099/ijs.0.034496-0
Fricke WF, McDermott PF, Mammel MK, Zhao S, Johnson TJ, Rasko DA, Fedorka-Cray PJ, Pedroso A, Whichard JM, Leclerc JE, White DG, Cebula TA, Ravel J: Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry . Appl Environ Microbiol 2009, 75: 5963–71. 10.1128/AEM.00786-09
Brinkkötter A, Klöss H, Alpert C, Lengeler JW: Pathways for the utilization of N-acetyl-galactosamine and galactosamine in Escherichia coli . Mol Microbiol 2000, 37: 125–35. 10.1046/j.1365-2958.2000.01969.x
Göker M, Garcáa-Blázquez G, Voglmayr H, Telleráa MT, Martán MP: Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora . PLoS ONE 2009, 4: e6319. 10.1371/journal.pone.0006319
Staley J, Krieg NR: Bacterial classification I. Classification of procaryotic organisms: an overview. In Bergey’s Manual of Systematic Bacteriology. First edition, Volume 1. Edited by: Krieg NR, Holt JG. Baltimore: The Williams & Wilkins Co; 1984:1–4.
Tindall BJ, Kampfer P, Euzeby JP, Oren A: Valid publication of names of prokaryotes according to the rules of nomenclature: past history and current practice. Int J Syst Evol Microbiol 2006, 56: 2715–20. 10.1099/ijs.0.64780-0
Lan R, Reeves P: Escherichia coli in disguise: molecular origins of Shigella . Microbes Infect 2002, 4: 1125–32. 10.1016/S1286-4579(02)01637-4
Acknowledgements
The authors gratefully acknowledge the help of Bettina Henze, DSMZ, for growing cells of DSM 30083T and of Susanne Schneider, DSMZ, for DNA extraction and quality control. Access to the record card for strain U5/41T provided by Flemming Scheutz of the Danish State Serum Institute is gratefully acknowledged. This work was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract no. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under contract no. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract no. DE-AC02-06NA25396, UT-Battelle and Oak Ridge National Laboratory under contract DE-AC05-00OR22725.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JPMK, CS and MG conducted the phylogenomic studies. JPMK, RLH, MG and HPK drafted the manuscript. JP, VM, CR and MR performed the laboratory experiments. AF, BF, LAG, OC, TR, AP, NNI, VM, NCK and TW sequenced, assembled and annotated the genome. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Meier-Kolthoff, J.P., Hahnke, R.L., Petersen, J. et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand in Genomic Sci 9, 2 (2014). https://doi.org/10.1186/1944-3277-9-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1944-3277-9-2