Abstract
Recent successes in the use of gene sequencing for patient care highlight the potential of genomic medicine. For genomics to become a part of usual care, pertinent elements of a patient’s genomic test must be communicated to the most appropriate care providers. Electronic health records may serve as a useful tool for storing and disseminating genomic data. Yet, the structure of existing EHRs and the nature of genomic data pose a number of pragmatic and ethical challenges in their integration. Through a review of the recent genome-EHR integration literature, we explore concrete examples of these challenges, categorized under four key questions: What data will we store? How will we store it? How will we use it? How will we protect it? We conclude that genome-EHR integration requires a rigorous, multi-faceted, and interdisciplinary approach of study. Problems facing the field are numerous, but few are intractable.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 2011, the National Human Genome Research Institute (NHGRI), released its new strategic plan in a landmark article titled Charting a Course for Genomic Medicine: From Base Pairs to Bedside. The manuscript highlighted the growing need for translational research to utilize genomic sequencing data for improvements in healthcare [1]. Translational science, in particular translational bioinformatics (TBI), seeks to unite biological data with patient care through a variety of innovative, systems-based approaches [2]. To support its shift in focus, the NHGRI created the Division of Genomic Medicine, strengthened collaborative, multi-institution translational projects, and reprioritized its funding [3]. This ongoing direction is driven by a growing number of reports of genome-driven diagnosis, treatment, and management. Researchers have used exome sequencing to identify the pathogenesis of refractory forms of disease and underlying genomic defects of patients with rare Mendelian phenotypes [4, 5]. Therapies targeted at tumors with genetic variants have become a mainstay of oncology treatment, while pharmacogenomics can be used to assess the likelihood of a patient’s response to common medications [6, 7]. Though these examples of the clinical applications of genomics have generated excitement among many key stakeholders, they are still disparate and often specialty specific. As the NHGRI’s strategic plan suggests, there are issues of operationalization and scalability that must be addressed in order for genomic medicine to become a part of comprehensive, standard patient care.
Perhaps the most pressing issue is the integration of patient genomic information into the electronic health record (EHR) [8]. Beginning in 2015, Meaningful Use regulations of the American Recovery and Reinvestment Act will penalize health care institutes that are not using EHRs for patient care [9]. Thus, a patient’s record will increasingly be used as a durable hub for diagnostic and treatment documentation and has been deemed a potentially convenient place to store his or her genomic data [10]. However, current EHR systems were not built for genomic data, and the characteristics of that data are still evolving, which pose multiple technical issues. Furthermore, there are numerous social and ethical consequences of capturing genomic information in EHR systems. For health care institutions interested in implementing EHR-driven genomic medicine, these concerns lead to four major questions: What data will we store? How will we store it? How will we use it? How will we protect it? Here, we elucidate both the pragmatic and ethical challenges of these questions based on a review of the current literature exploring the integration of the genome and the EHR.
The Current Landscape of Genome-EHR Endeavors
Recent publications from academic centers either in the NHGRI’s multi-institutional collaborations or independently exploring genomic medicine have laid the groundwork for the integration of genomics in the EHR. The NHGRI founded the Electronic Medical Records and Genomics (eMERGE) network in 2007 to explore how EHR data could be juxtaposed with large DNA repositories in GWAS studies to identify the phenotypic importance of novel variants [11]. Although, the network initially emphasized using EHR-based data for discovery of disease-associated variants, its second phase, initiated in 2011, is primarily concerned with genome-EHR integration. Similarly, the Clinical Sequencing and Exploratory Research (CSER) consortium was created in 2010 by the NHGRI to develop methods for incorporating large-scale sequencing data into clinical care and to study the ethical, legal, and social implications (ELSI) of the process [12]. Though each CSER site focuses on a different medical application of genomics, most sites are concerned with the use of EHRs for genomic medicine. In sum, there are 9 eMERGE sites and 18 CSER sites (Table 1).
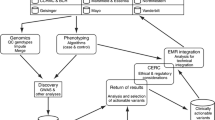
Among the eMERGE sites, Mayo Clinic [13], Mount Sinai School of Medicine [14, 15], and Vanderbilt [16] have written about their experiences of incorporating pharmacogenomics data into their EHR systems. Sites independent of eMERGE and CSER such as St. Jude Medical Center [17, 18], Harvard University [19], the University of Chicago [20], the University of Maryland [21], and the NIH’s care center have discussed similar ventures [22]. These publications rarely report outcomes for particular interventions, but instead share valuable institutional experiences in genome-EHR integration. Indeed, based on the recent genome-EHR publications, a generalized prototypic workflow tracing the steps from genetic test to EHR-based clinical application can be extrapolated and is described in Fig. 1. In addition, the publications detail where challenges arise as genomic data is introduced into EHR systems and will be referenced accordingly throughout this review.
Prototypic workflow of genetic testing to EHR data. Any high throughput sequencing test will identify thousands of variants, genomic sites where the patient’s sequence is different from the human genome reference sequence. Variants are identified and classified by pathogenicity in a process called annotation. Semi-automated bioinformatics tools allow the clinical laboratory geneticist to remove low-quality and common variants and flag variants with known pathogenicity. The laboratory geneticist then reviews the remaining, uncommon variants, with a review of the literature or additional genomic databases. Annotation and prediction of phenotypic impact is a straightforward process if the variant is well classified, but if the variant is novel, a variant of unknown significance (VUS), or there is conflicting information from different sources, annotation may require extensive investigation by individuals or panels of experts. The results of genomic testing describing the variant and clinical significance are then disseminated to providers and, eventually, patients through a variety of means
The first issues arise early in the workflow, as genomic data is generated, annotated, and deemed actionable.
What Data to Store?
Due to the complex nature of genomic data and the evolving literature base of genomic medicine, identifying the information that will be integrated into the EHR poses a number of challenges. In order to choose the data to be captured and stored, institutions must differentiate the types of data generated by various genetic tests, establish variant pathogenicity through consistent annotation, and determine the clinical relevance of specific variants.
Types of Genetic Tests
While Whole Genome/Exome Sequencing (WGS/WES) may eventually become standard forms of clinical testing, the massive amount of information generated poses data management challenges. Currently, most clinical genetic tests rely on targeted testing of components of an individual’s genome. These tests are generally faster and less expensive than WGS/WES, but require a priori identification of the gene or variant of interest. WGS/WES tests, conversely, are more comprehensive and can identify novel variants. They may also be done for specific purposes, such as identifying a hereditary cause for a patient’s colon cancer, but will likely be accompanied by additional variants with a phenotypic impact. These auxiliary results have been called incidental findings (IF). Decisions must be made about whether and how to return IFs to patients as they may cause potential confusion and/or stress. Given the large number of results that could potentially be returned to the patient, there is concern that incidental findings of lesser importance could distract the patient from results of greater impact. Some institutions allow patients prior to testing to opt out of receiving incidental findings; others have used a staged release of results to allow patients to focus on the most important findings first [23]. Institutions should consider their capacities and resources to manage IFs and the large swath of data generated by WGS/WES when choosing genomic tests.
Another important consideration for test choices is the source of tissue. During a patient’s lifetime, they may have multiple sequencing tests performed for different purposes on different tissues, such as tumor testing for somatic mutations. Data management complications arise in that what is incidental in one setting may be intentional in another. Furthermore, genomic variants from unique tissues may require independent representation in EHR systems. As the cost of genotyping drops and large-scale genomic testing becomes more common, a major question in representing the genome in the EHR may be, “Which genome?” [24]
Annotation
Once a genomic test is chosen and sequencing data is generated, institutions must determine which elements of the data are going to be stored in the EHR. Currently, the parts of the genome that are most relevant to patient care are the variants that predispose an individual to a pathogenic or abnormal phenotype. Yet, accurately identifying, classifying, and ranking the most actionable variants in the annotation process is by no means straightforward.
A consistent annotation pipeline might help identify the potential pathogenicity of variants, but the process is difficult due to a plethora of disconnected variant databases. Multiple variant databases exist, but each carries different types of information, with no authoritative resource for annotation and determination of a variant’s clinical relevance [25]. Larger, popular databases used for annotation, such as Human Gene Mutation Database (HGMD) [26], lack medically applicable phenotypic information, while newer clinically oriented variant databases, such as GeneReviews and ClinVar, are currently limited in size. A survey of CSER sites found that institutions used many different databases for annotation [27]. The heterogeneity of resources increases the possibility that for any given variant, sites may reach inconsistent conclusions about pathogenicity and clinical implications. CSER and eMERGE leaders have long called for a well-curated, open-access, and clinically relevant variant database to simplify and standardize the annotation process; [28] the NHGRI has responded accordingly by creating an RFP for the creation of such a database [29]. An ideal solution would involve extensive sharing of information from different databases and clinical laboratories [30]. However, this solution might raise concerns from individuals who do not want their information shared and from companies claiming proprietary genetic data. Even if a variant is determined to be clinically actionable by the testing laboratory, treating physicians must still decide which clinical to take, if any.
Clinical Relevance
Currently, decisions about how a variant should be dealt with in the medical setting are determined by institution-based expert panels. For certain genomic applications, such as targeted tumor therapy, the clinical response to a handful of known variants has a substantial literature base and is straightforward. However, for other medical uses of genomics, in particular pharmacogenomics, the evidence for action is more sparse. The panels must sift through the various forms of evidence and, as with the annotation process, their conclusions may differ between centers [27]. Some of the aforementioned variant databases, such as ClinVar or PharmGKB, have clinical guidelines for certain variants, but health care institutions may be wary to rely solely on these for genomic recommendations. Accountability is a major issue, as hospitals and clinics may be concerned about liability for poor outcomes based on novel applications of genomics to patient care. Furthermore, characteristics of genomic tests make them subject to genetic exceptionalism, in which they are held to different standards than other medical tests [31, 32]. The problem is exacerbated by the limited and fractured evidence base for many clinical genomic interventions.
The University of Maryland genome-EHR integration project summarizes the difficulty in establishing a solid foundation of literature-based evidence for their creation of pharmacogenomics decision support for the anti-platelet agent, clopidogrel [21]. Smaller and non-randomized studies have given preliminary evidence in favor of genotype-driven anti-platelet management and guidelines offered by the Clinical Pharmacogenetics Implementation Consortium (CPIC), a project of PharmGKB and the Pharmacogenomics Research Network (PGRN), advocate altered therapies for poor clopidogrel metabolizers. However, due to the low rate of adverse events currently associated with clopidogrel, the UM team notes that powering a randomized control trial (RCT) to test the benefits of genotyping would require thousands of subjects; no trials to date have offered such evidence. Thus, professional societies such as the American College of Cardiology are cautious about advocating routine genomic testing for anti-platelet therapy without stronger evidence [33]. Not only are few guidelines available to direct genomic care, but those that exist may be in conflict.
A recent series of randomized controlled trials comparing the impact of genotype-driven warfarin dosing on the time spent in a therapeutic range exemplified how the clinical relevance of genomic testing remains a contentious issue in the broader medical literature. One trial showed mild improvements with intervention; [34] two showed no statistically significant differences between groups [35, 36]. The publications prompted PharmGKB and CPIC to post an alert on their warfarin web guidelines stating that the papers were under evaluation for their impact on existing recommendations. Many readers found the trials to be substantial evidence against warfarin genetic testing. However, the studies were criticized because control subjects had an unusual high frequency of International Normalized Ratio (INR) testing, the INR measurements were only short term, and algorithms compared were complex beyond the genotype variables [37].
It may be some time before a strengthened, consistent literature base for genomic medicine interventions exists. The sheer number of putatively actionable genomic variants will preclude prospective RCT examining outcomes of interventions for each one [38]. Clinical actions for such variants may need to rely on alternative forms of evidence, such as family concordance, evolutionary conservation, linkage analysis, and functional testing [39]. Skeptics may question the strength of such evidence for usual care, as well as the role of genotype-driven medicine when it is contextualized with non-genomic alternatives. For example, one might consider RCTs for genotying of clopidogrel to be a poor use of resources given that new anti-platelet agents, ticagrelor and prasugrel, are less dependent on a patient’s genetics for bioactivation [40]. Thus, support for a firm base of clinical trials to evaluate genomic interventions may be difficult to find in the near future.
As resources to support the annotation processes become more consistent and best practices are established to clarify genomic sequencing’s place in medical care, the next question for institutions will be how to store actionable variants.
How to Store?
All the individual sequences that are aligned to create an individual’s entire genome sequence can contain approximately 90 gigabytes of data and has about 3.5 million variants [41]. Current EHR systems are not structured to carry, analyze, or manage raw genomic data; consequently, sequencing data is not stored directly in the EHRs. Instead, specific clinical laboratory results are currently captured in semi-structured, free-text documents written by geneticists or laboratory physicians [27]. These documents are usually sent to a key care provider or stored in the EHR as a PDF file. Unfortunately, these formats are not machine-readable and cannot be used by EHRs to generate clinical decision support tools. Although the reports are technically in the EHR, providers may not be aware of them—especially for care that does not occur soon after genomic testing.
Many institutions exploring genome-EHR integration leverage existing discrete laboratory test result systems to make parts of the genomic data machine-readable, but this solution poses some problems. First, there is no EHR-supported and accepted structure of standard variant identifiers. Following variant naming standards established by the Human Genome Variation Society (HGVS) is required by most journals and databases. However, “traditional” nomenclature systems defined by early publications have been carried over in the literature and are used by many laboratories for certain variants, resulting in inconsistent referencing of variants [42, 43]. Even if the genomics community fully committed to the HGVS nomenclature, the intricacy of the genome may preclude a fully computable naming system that would simultaneously provide unique identifiers for every possible variant. Without a standard way to reference a patient’s variant in the EHR, systematic storage and retrieval of genomic data becomes difficult. Goldspiel et al. [22] note such difficulties in their system when the naming for an allele changed from HLAB5701 to HLAB57:01. Even such minor discrepancies can have major impacts on clinical computing systems. Complex and comprehensive lookup libraries correlating multiple genomic identifiers between multiple laboratory and EHR result fields may be required for effective genomic storage and reporting.
A further difficulty with storing discrete genomic results in the EHR is the choice in granularity of the test result that must be made. For example, on the coarsest level, a discrete report for the drug-metabolizing enzyme CYP2C19 could state in binary terms whether or not the patient is an abnormal CYP2C19 metabolizer. However, an “abnormal” metabolizer could metabolize drugs too slowly or too quickly, making a binary indicator insufficient. Instead, the report might indicate the patient is a heterozygote for CYP2C19*3/*2 variants or heterozygote for variants rs1295183 and rs1384513. While these latter formats can represent different impacts of specific variants, they are less human readable and more challenging to maintain. Such granular lab results have the potential to drastically increase the number of defined elements in libraries of result definitions, rules engines, and interface engines. Though granularity does not appear to drastically increase processing time for rules engines [44], it significantly amplifies the number and complexity of reports that must be curated, tested, and maintained.
Regardless of how the discrete lab report nomenclature is structured, there is an additional choice to be made regarding which variants will be captured and a resulting loss of data. The rest of the genomic data must then be stored elsewhere or abandoned completely. Yet, clinical genomics research is rapidly evolving; it is likely that data filtered today as clinically irrelevant may become relevant in the future [45]. An interdisciplinary group of experts at the workshop on ‘‘Integration of Genetic Test Results into Electronic Health Records’’ convened by the National Heart Lung and Blood Institute stated as one of the desiderata of genome-EHR integration that raw genomic information be kept fully intact and accessible to avoid this problem [46]. Such an aspiration is not feasible using discrete reports alone for storage of the genome in the EHR. The workshop experts suggested lossless compression of sequencing data to reduce file size. Alternatively, the genomic data could be stored in databases separate from the EHR that can be accessed as needed: Pulley et al. [16] discuss the “sequestration” of their raw sequencing data deemed irrelevant at the time of their study into a secure database.
Given that an institution with unlimited resources could determine which aspects of which genome to store and how it wants to store them, further questions arise in regard to how such information will be retrieved and utilized.
How to Use the Data?
Targeted dissemination of clinically relevant, patient-specific data to health care providers is a fundamental step of scalable genomic medicine and EHR-based clinical decision support (CDS) may be an important way to provide it [47, 48]. CDS refers to information made available to health care providers at different points in care processes to strengthen their decision-making. Certain forms of CDS have been shown to be an effective way to alter processes in care delivery, as well as to improve functional and clinical outcomes for a wide range of health care activities [49]. While there are many potential benefits of disseminating genomic data to providers through EHR-based decision support tools, there are challenges to successful implementation of these tools.
CDS is a broad term that includes many types of interventions; [50] it is unclear which type is best suited for genomic information. Indeed, in many ongoing pharmacogenomics projects, a shotgun approach is being used to disseminate the data in as many forms as possible [14]. Genomic data could potentially take advantage of decision support tools that are both passive, where tools support a physician seeking a response to a question, and active, where tools anticipate the information a physician will need and present it. Existing examples of genomic-CDS include static PDF reports, lines in the patient summary, statements in the problem list, separate genomics “tabs” in the EHR, and drug-gene alert “pop-ups” triggered within a computerized physician order entry (CPOE) system.
Drug-gene alerts are an attractive form of pharmacogenomic CDS as they may leverage existing drug–drug interaction rules engines and serve as a simple platform for “pre-emptive” genomics where genotyping is done prior to and without knowledge of the specific genetic event [15]. For example, reactive genotyping would include ordering a HLA*B5701 test for a patient diagnosed with HIV prior to initiation of abacavir to avoid a hypersensitivity reaction; pre-emptive genotyping would involve a healthy patient having WES and discovering the HLA*B5701 as an incidental finding. The pre-emptive data would only be of use if the patient were to be prescribed abacavir in the future. Genome-wide clinical data might bypass the need for multiple expensive reactive tests with slow turnaround times that disrupt the clinical workflow [21]. Because pre-emptive genomic testing may be temporally separated from clinical utilization, CDS solutions that ensure a provider is notified at the appropriate time are required.
Most EHR systems already have CPOE-based alerts; however, they are far from a silver bullet as they carry with them a number of challenges such as the threat of “alert fatigue” [51–53]. Key elements of effective alerts have been identified including consistent design elements, appropriate visuals, supporting advice, and clinical relevance [54]. However, the research base of best practices for genomic-CDS visual and educational content is limited [55]. And, as stated above, stable, evidence-based clinical relevance is still a major challenge for genomics. Furthermore, clinical relevance for decision support tools not only refers to available guidelines, but also to the tool’s ability to accurately address the information needs of a specific clinical event between patient and provider.
Goldspiel et al. [22] highlight the difference between pre- and post-genomic testing alerts, exemplifying the need for alerts that are relevant to an institution’s patient and provider populations. Their CDS system includes alerts that fire when a provider prescribes the HIV medication, abacavir, for a patient. Pre-test alerts inform the provider of the risk for hypersensitivity reactions among patients with HLA*B5701 variants and recommend genetic testing prior to initiation of treatment. The post-test alerts draw on a patient’s genomic data to inform the provider that the patient has such a variant and that alternative treatment should be pursued. Providers overrode 100 % of their pre-test abacavir alerts [22]. The authors note that HLA testing is routine and that all of the patients had received the tests elsewhere or were already taking abacavir, thus making it reasonable for the providers to override. In this particular setting, using post-test alerts would likely prove more useful for the institution’s providers and also reduce the total number of alerts to which they are subjected. The example highlights the need to consider both when and how an alert is intended to fire, which must be recognized for each specific drug–gene interaction of interest and each institution’s particular patient population. Furthermore, alert timing is only one element of an alert system’s clinical relevance.
Another aspect of clinical relevance for alerts is the specificity they have for firing in appropriate clinical events. In the above example, the pre-test abacavir alerts have low specificity because they appear frequently in situations in which they are irrelevant. An alert with higher specificity would only fire for patients who weren’t already taking abacavir and who had not had HLA testing. Rules engines could apply this information to algorithms that determine when alerts fire. However, such information is rarely available in machine-readable format in current EHR systems. Thus, one of the primary hurdles for increasing the specificity, and thus efficacy, of genomic alert systems is the limited, usable data in EHR systems in general.
Given the challenges of active CDS for genomic purposes, it is not unreasonable to look for passive or “semi-active” solutions. O’donnell et al. [20] describe their passive CDS system as a provider portal that contains all of a patient’s pharmacogenomic information and utilizes color-coded traffic light icons to signify different levels of priority. Hoffman et al. [18] also describe the creation of a passive pharmacogenomics tab in their EHR system, which functions like a traditional lab results tab but with a “lifetime nature” to capture the temporal aspects. Finally, many groups found some genomic drug information worthy of the patient’s problem list. Bell et al. [17] even utilized the problem list entry as the discrete data for active CDS tools, bypassing issues of multiple lab results contributing to one phenotype (e.g., CYP2C9 and VKOR both impacting warfarin metabolism).
Whether an institution chooses to use passive, active, or both forms of CDS, the uptake and usage of the tools depend largely on provider acceptance. Successful CDS require that providers trust the tools as a source of support and that they understand how the tools can improve their practices [56]. The knowledge generated by genomics already exceeds the mental capacity of any given physician; thus, it will be difficult for individual providers to assess the reasoning behind each institutional CDS tool. When a provider must abdicate her decisions to a computer, the “black box” effect can threaten her confidence in her actions. CDS developers can address this issue by stating the sources for decision support recommendations and offering the educational resources needed for personal investigation whenever possible. Genomic-CDS projects have sought to educate providers about their interventions prior to exposure [14, 22], upon the launch of a new drug–gene interaction through newsletters and presentations [18], or within the CDS itself. Mayo Clinic, for example, provides education within the CDS by linking to short summaries in the AskMayoExpert tool [13]. Offering such resources through active CDS ensures that providers have educational resources at the point-of-care [47], but providers’ exact knowledge gaps regarding genomic medicine remain unclear.
Myriad elements go into the creation of effective CDS. As the issue of alert fatigue suggests, poorly designed, maintained, and executed CDS are not only ineffective, but potentially harmful. While a sizeable pool of research has begun to reveal best practices for particular CDS uses, such as the prevention of drug–drug interactions [57, 58], there is currently no consensus on which forms of decision support are most appropriate for genomic data [59]. Exploratory efforts of genome-EHR integration will begin to establish genomic CDS best practice evidence. The additional complexities of sequencing data will undoubtedly require CDS tools that are uniquely adapted for genomic medicine. Creative approaches to CDS will likely be required as the subtleties of CDS design may influence their cognitive impact on providers [54, 60]. Further insight from non-medical fields may aid in the development of effective genomic CDS [61].
Genomic data in the EHR will not only be accessible to providers, but patients as well. Given the static nature of the stored results, an important concern for institutions will be how to disseminate pertinent genetic information to patients in secure ways at appropriate times.
How to Protect the Patient? Return of Results and Privacy
Returning test results to patients is a key ethical concern for genomics in general, beyond EHR integration. Given the complexities and implications of genomic test results, it is important that they be communicated to patients with sufficient educational support. Indeed, the American College of Medical Genetics and Genomics (ACMG) issued a recommendation in 2013 that patients be educated both before and after the testing [62]. It has been noted that, despite the growing ability for automated annotation and development of comprehensive knowledge bases, the role of the care provider in interpreting and conveying genomic information remains of paramount importance [63]. Ideally, results could be returned with in-person explanation from a genetic counselor, supported by mixed-media educational tools. Yet, there will likely be insufficient genetic counselors to facilitate all such conversations. The task of explaining genomic test results will then fall to others. Genetic counseling is a specialized skill that requires experience and practice, yet surveys suggest that genetic training for primary care providers is limited [64]. If all care providers may be responsible for returning results, some genetic counseling skills would need to become a component of medical training. Given the need for successful genomic CDS tools to have supporting educational information, it will be important to tailor the tools to the point-of-care needs of the different types of physicians [65]. Providers will further need to learn their legal responsibilities to deal with both intended and incidental findings [66].
Regardless of who ultimately returns the results, best practices will need to be established about when the results are given. For targeted and reactive genomic tests, it is reasonable to return the results as soon as they are available. However, for pre-emptive tests or incidental findings, it may make more sense to inform the patient that he or she has the finding when it becomes clinically relevant. For instance, if a patient is found to have a variant in TPMT that alters his metabolism of thiopurine, it may only be important to discuss the implications of the result if the patient needs such medications. This course of action, however, would be dependent on robust EHR and CDS systems being in place. Such situations are particularly important for pediatric populations [67, 68]. Additionally, situations may arise in which a finding that was once deemed clinically irrelevant is discovered to be pathogenic. Re-contacting patients raises a number of additional questions about what updates are worth informing the patient and whose responsibility it is to reach the patient [69]. As storage of genomic data in the EHR likely occurs soon after the tests are performed, protocols and systems will be needed to help providers deliver the information at the most appropriate times and to ensure that data is appropriately transferred to new EHR systems if the patient changes care providers.
One of the potential stressors that patients may face upon the return of their results is a concern about who else should know about the findings. The social implications of genetic findings have long been an ethical topic of concern, as an individual’s genetic pattern may reveal information about family members or communities [70]. These issues remain true for genomic test data stored within EHR systems. The Genetic Information Nondiscrimination Act (GINA) of 2008 prevents health insurers and employers from discriminating based on genetic information. However, the act does not seek to protect the privacy of genetic information. The privacy of genetic information in the EHR is protected under HIPAA legislation that covers all medical information; there are currently no national provisions for additional protection of genetic information as it is not clear that additional protections would be beneficial, although several states have passed legislation specifically protecting genetic information [71, 72].
Genomic information embedded in the EHR could serve as an important source for both research data and institutional quality assurance and improvement, especially for institutions striving to become Rapidly Learning Health systems [73]. Use of that data for research should be approached with the utmost concern for patient privacy, while policies should take care not to unnecessarily limit clinical quality improvement activities that benefit all patients. New approaches to informed consent and de-identification must be developed to accommodate the potential for future use of digitally stored genetic data. The need for such approaches was highlighted by the legal battle of the Havapusai Native Americans [74], when DNA samples in biobanks were used for research purposes other than those to which they had consented. The research exposed information about the tribe that conflicted with its cultural beliefs as well as increased the risks of stigmatization about mental health issues. On the other hand, routine inclusion of de-identified information about rare variants into public databases will be necessary for meaningful clinical interpretation of these variants. It will be important to ensure that patients are adequately informed about how their genomic data will be used and ensure robust de-identification of genetic data used for quality improvement. Strategies for consenting patients to additional research, such as tiered consenting or presumed consent, may need to be implemented [74].
Conclusion
Since the NHGRI’s pivotal shift toward the use of genomic data in medical practice, a vast amount has been discovered about the process. In parallel, the EHR has become an important facet in bridging laboratory sequencing results to personalized patient care. Efforts by members of CSER, EMERGE, and others have provided a fruitful source of experiential knowledge about the integration of genomics and EHRs. In addition to the many anticipated benefits of genomic medicine, these projects have illuminated the pitfalls and challenges in the acquisition, representation and, retrieval of genomic data. These challenges revolve around issues of shared knowledge resources, nomenclature and identification, data storage, clinical decision support, return of results, and privacy protection. In this article, we have attempted to outline these issues and provide concrete examples from the literature. Though we have not addressed them here, we recognize that other aspects of genomic-EHR integration, such as the impact of epigenomics [75], the just distribution of resources, and the influence of different EHR systems [30], are of great concern and deserve attention. The field of genomic medicine is vast and requires a rigorous, multi-faceted, and interdisciplinary approach of study. We look forward to the solutions that will be developed for the challenges outlined in this review.
References
Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470(7333):204–13. doi:10.1038/nature09764.
Sarkar IN, Butte AJ, Lussier Ya, Tarczy-Hornoch P, Ohno-Machado L. Translational bioinformatics: linking knowledge across biological and clinical realms. J Am Med Inf Assoc. 2011;18(4):354–7. doi:10.1136/amiajnl-2011-000245.
NHGRI. National Human Genome Research Institute FY 2014 Budget. Bethesda; 2014:1–28.
Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13(3):255–62. doi:10.1097/GIM.0b013e3182088158.
Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;369(16):1502–11. doi:10.1056/NEJMoa1306555.
Stegmeier F, Warmuth M, Sellers WR, Dorsch M. Targeted cancer therapies in the twenty-first century: lessons from imatinib. Clin Pharmacol Ther. 2010;87(5):543–52. doi:10.1038/clpt.2009.297.
Miao J, Liu R, Li Z. Cytochrome P-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360(21):2250–1.
Hoffman MA. The genome-enabled electronic medical record. J Biomed Inf. 2007;40(1):44–6. doi:10.1016/j.jbi.2006.02.010.
Medicaid C for M and Payment Adjustments & Hardship Exceptions Tipsheet for Eligible Professionals Payment Adjustments for Medicare Eligible Professionals. 2014:1–5. http://www.cms.gov/Regulations-and-Guidance/Legislation/EHRIncentivePrograms/Downloads/PaymentAdj_HardshipExcepTipSheetforEP.pdf.
Overby CL, Tarczy-Hornoch P, Hoath JI, Kalet IJ, Veenstra DL. Feasibility of incorporating genomic knowledge into electronic medical records for pharmacogenomic clinical decision support. BMC Bioinformatics. 2010;11(Suppl 9):S10. doi:10.1186/1471-2105-11-S9-S10.
Gottesman O, Kuivaniemi H, Tromp G, et al. The electronic medical records and genomics (eMERGE) network: past, present, and future. Genet Med. 2013;15(10):761–71. doi:10.1038/gim.2013.72.
Clinical Sequencing Exploratory Research (CSER). Natl Hum Genome Res Inst. 2014. http://www.genome.gov/27546194.
Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89(1):25–33. doi:10.1016/j.mayocp.2013.10.021.
Gottesman O, Scott S. The CLIPMERGE PGx program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther. 2013;94(2):214–7. doi:10.1038/clpt.2013.72.The.
Scott S. Clinical pharmacogenomics: opportunities and challenges at point-of-care. Clin Pharmacol Ther. 2013;93(1):33–5. doi:10.1038/clpt.2012.196.Clinical.
Pulley JM, Denny JC, Peterson JF, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92(1):87–95. doi:10.1038/clpt.2011.371.
Bell GC, Crews KR, Wilkinson MR, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inf Assoc. 2014;21(e1):e93–9. doi:10.1136/amiajnl-2013-001993.
Hoffman JM, Haidar CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet. 2014;. doi:10.1002/ajmg.c.31391.
Fusaro V, Brownstein C. Development of a Scalable Pharmacogenomic Clinical Decision Support Service. AMIA Jt Summits Transl Sci Proc. 2013: 60. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3814487/. Accessed 10 June 2014.
O’Donnell PH, Danahey K, Jacobs M, et al. Adoption of a clinical pharmacogenomics implementation program during outpatient care-initial results of the University of Chicago “1,200 Patients Project”. Am J Med Genet C Semin Med Genet. 2014;. doi:10.1002/ajmg.c.31385.
Shuldiner AR, Palmer K, Pakyz RE, et al. Implementation of pharmacogenetics: the University of Maryland personalized anti-platelet pharmacogenetics program. Am J Med Genet C Semin Med Genet. 2014;. doi:10.1002/ajmg.c.31396.
Goldspiel BR, Flegel WA, Dipatrizio G, et al. Integrating pharmacogenetic information and clinical decision support into the electronic health record. J Am Med Inf Assoc. 2013;21:522–8. doi:10.1136/amiajnl-2013-001873.
Kullo IJ, Haddad R, Prows CA, et al. Return of results in the genomic medicine projects of the eMERGE network. Front Genet. 2014;5:50. doi:10.3389/fgene.2014.00050.
Leary R, Sausen M. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4(162):1–21. doi:10.1126/scitranslmed.3004742.Detection.
Johnston JJ, Biesecker LG. Databases of genomic variation and phenotypes: existing resources and future needs. Hum Mol Genet. 2013;22(R1):R27–31. doi:10.1093/hmg/ddt384.
Dorschner MO, Amendola LM, Turner EH, et al. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet. 2013;93(4):631–40. doi:10.1016/j.ajhg.2013.08.006.
Tarczy-Hornoch P, Amendola L. A survey of informatics approaches to whole-exome and whole-genome clinical reporting in the electronic health record. Genet Med. 2012;2013(15):824–32. doi:10.1038/gim.2013.120.A.
Chute CG, Kohane IS. Genomic medicine, health information technology, and patient care. JAMA. 2013;309(14):1467–8.
NHGRI. Clinically Relevant Genetic Variants Resource: A Unified Approach for Identifying Genetic Variants for Clinical Use (U01). 2012. http://grants.nih.gov/grants/guide/rfa-files/RFA-HG-12-016.html.
Peterson J, Bowton E, Field J. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet Med. 2013;15(10):833–41. doi:10.1038/gim.2013.109.Electronic.
Green M, Botkin J. “Genetic exceptionalism” in medicine: clarifying the differences between genetic and nongenetic tests. Ann Intern Med. 2003;(32):571–576. http://annals.org/article.aspx?articleid=716276. Accessed 29 May 2014.
Evans JP, Burke W, Khoury M. The rules remain the same for genomic medicine: the case against “reverse genetic exceptionalism”. Genet Med. 2010;12(6):342–3. doi:10.1097/GIM.0b013e3181deb308.
Angiolillo D, Ferreiro J, Price M, Kirtane A, Stone G. Platelet function and genetic testing. J Am Coll Cardiol. 2013;62(17):S21–31.
Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med. 2013;369(24):2294–303. doi:10.1056/NEJMoa1311386.
Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369(24):2283–93. doi:10.1056/NEJMoa1310669.
Verhoef TI, Ragia G, de Boer A, et al. A randomized trial of genotype-guided dosing of acenocoumarol and phenprocoumon. N Engl J Med. 2013;369(24):2304–12. doi:10.1056/NEJMoa1311388.
Zineh I, Pacanowski M, Woodcock J. Pharmacogenetics and coumarin dosing–recalibrating expectations. N Engl J Med. 2013;369:2273–5.
Shirts BH, Jacobson A, Jarvik GP, Browning BL. Large numbers of individuals are required to classify and define risk for rare variants in known cancer risk genes. Genet Med. 2013;16(7):529–34. doi:10.1038/gim.2013.187.
Richards CS, Bale S, Bellissimo DB, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med. 2008;10(4):294–300. doi:10.1097/GIM.0b013e31816b5cae.
Capodanno D, Ferreiro JL, Angiolillo DJ. Antiplatelet therapy: new pharmacological agents and changing paradigms. J Thromb Haemost. 2013;11(Suppl 1):316–29. doi:10.1111/jth.12219.
Lupski JR, Reid JG, Gonzaga-Jauregui C, et al. Whole-genome sequencing in a patient with Charcot–Marie–Tooth neuropathy. N Engl J Med. 2010;362(13):1181–91. doi:10.1056/NEJMoa0908094.
Pandey KR, Maden N, Poudel B, Pradhananga S, Sharma AK. The curation of genetic variants: difficulties and possible solutions. Genomics Proteomics Bioinform. 2012;10(6):317–25. doi:10.1016/j.gpb.2012.06.006.
Berwouts S, Morris MA, Girodon E, Schwarz M, Stuhrmann M, Dequeker E. Mutation nomenclature in practice: findings and recommendations from the cystic fibrosis external quality assessment scheme. Hum Mutat. 2011;32(11):1197–203. doi:10.1002/humu.21569.
Deshmukh VG, Hoffman MA, Arnoldi C, Bray BE, Mitchell JA. Efficiency of CYP2C9 genetic test representation for automated pharmacogenetic decision support. Methods Inf Med. 2009;48(3):282–90. doi:10.3414/ME0570.
Shirts BH, Parker LS. Changing interpretations, stable genes: responsibilities of patients, professionals, and policy makers in the clinical interpretation of complex genetic information. Genet Med. 2008;10(11):778–83. doi:10.1097/GIM.0b013e31818bb38f.
Masys DR, Jarvik GP, Abernethy NF, et al. Technical desiderata for the integration of genomic data into Electronic Health Records. J Biomed Inf. 2012;45(3):419–22. doi:10.1016/j.jbi.2011.12.005.
Overby C, Kohane I, Kannry J. Opportunities for genomic clinical decision support interventions. Genet Med. 2013;15(10):1–13. doi:10.1038/gim.2013.128.Opportunities.
Overby C, Tarczy-Hornoch P. Personalized medicine: challenges and opportunities for translational bioinformatics. Per Med. 2013;10(5):453–462. http://www.futuremedicine.com/doi/abs/10.2217/pme.13.30. Accessed 29 May 2014.
Bright T, Wong A, Dhurjati R. Effect of clinical decision-support systems: a systematic review. Ann Intern Med. 2012;157(1):29–43.
Richardson J, Ash J, Sittig D. Multiple perspectives on the meaning of clinical decision support. AMIA Annu Symp Proc. 2010;(8):1427–1431.
Carspecken CW, Sharek PJ, Longhurst C, Pageler NM. A clinical case of electronic health record drug alert fatigue: consequences for patient outcome. Pediatrics. 2013;131(6):e1970–3. doi:10.1542/peds.2012-3252.
Van Der Sijs H, Aarts J, Vulto A, Berg M. Overriding of drug safety alerts in computerized physician order entry. J Am Med Inform Assoc. 2006;13(2):138–48. doi:10.1197/jamia.M1809.Computerized.
Ash J, Sittig D, Poon E. The extent and importance of unintended consequences related to computerized provider order entry. J Am Med Inform Assoc. 2007;14(4):415–23. doi:10.1197/jamia.M2373.Introduction.
Horsky J, Schiff GD, Johnston D, Mercincavage L, Bell D, Middleton B. Interface design principles for usable decision support: a targeted review of best practices for clinical prescribing interventions. J Biomed Inform. 2012;45(6):1202–16. doi:10.1016/j.jbi.2012.09.002.
Welch BM, Kawamoto K. Clinical decision support for genetically guided personalized medicine: a systematic review. J Am Med Inform Assoc. 2012;20(2):388–400. doi:10.1136/amiajnl-2012-000892.
Hartzler A, McCarty C. Stakeholder engagement: a key component of integrating genomic information into electronic health records. Genet Med. 2013;15(10):792–801. doi:10.1038/gim.2013.127.Stakeholder.
Phansalkar S, Desai A, Choksi A, et al. Criteria for assessing high-priority drug–drug interactions for clinical decision support in electronic health records. BMC Med Inform Decis Mak. 2013;13(1):65. doi:10.1186/1472-6947-13-65.
Jung M, Riedmann D, Hackl WO, et al. Physicians’ Perceptions on the usefulness of contextual information for prioritizing and presenting alerts in computerized physician order entry systems. BMC Med Inform Decis Mak. 2012;12(1):111. doi:10.1186/1472-6947-12-111.
Welch BM, Kawamoto K. Clinical decision support for genetically guided personalized medicine: a systematic review. J Am Med Inform Assoc. 2013;20:388–400. doi:10.1136/amiajnl-2012-000892.
Horsky J, Zhang J, Patel VL. To err is not entirely human: complex technology and user cognition. J Biomed Inform. 2005;38(4):264–6. doi:10.1016/j.jbi.2005.05.002.
Wu HW, Davis PK, Bell DS. Advancing clinical decision support using lessons from outside of healthcare: an interdisciplinary systematic review. BMC Med Inform Decis Mak. 2012;12(1):90. doi:10.1186/1472-6947-12-90.
Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–74. doi:10.1038/gim.2013.73.
McGuire A, McCullough L, Evans J. The indispensable role of professional judgment in genomic medicine. JAMA. 2013;309(14):1465–6. doi:10.1001/jama.2013.1438.The.
Houwink EJ, van Luijk SJ, Henneman L, van der Vleuten C, Jan Dinant G, Cornel MC. Genetic educational needs and the role of genetics in primary care: a focus group study with multiple perspectives. BMC Fam Pract. 2011;12(1):5. doi:10.1186/1471-2296-12-5.
Haga SB, Tindall G, O’Daniel JM. Professional perspectives about pharmacogenetic testing and managing ancillary findings. Genet Test Mol Biomark. 2012;16(1):21–4. doi:10.1089/gtmb.2011.0045.
Clayton E, Haga S, Kuszler P. Managing incidental genomic findings: legal obligations of clinicians. Genet Med. 2013;15(8):624–9. doi:10.1038/gim.2013.7.Managing.
College A. Incidental findings in clinical genomics: a clarification. Genet Med. 2013;15(8):664–6. doi:10.1038/gim.2013.82.
Burke W, Matheny Antommaria AH, Bennett R, et al. Recommendations for returning genomic incidental findings? We need to talk! Genet Med. 2013;15(11):854–9. doi:10.1038/gim.2013.113.
Pyeritz R. The coming explosion in genetic testing: is there a duty to recontact. N Engl J Med. 2011;365:1367–9.
Beskow LM, Burke W, Merz JF, et al. Informed consent for population-based research involving genetics. JAMA. 2001;286(18):2315–21.
Table of State Statutes Related to Genomics. Natl Hum Genome Res Inst. 2014. http://www.genome.gov/27552194. Accessed 10 June 2014.
Sarata A, Jones N, Staman J. Genetic Information Nondiscrimination Act of 2008 and the Patient Protection and Affordable Care Act of 2010: Overview and legal analysis of potential. CRS Rep Congr. 2010. http://scholar.google.com/scholar?hl=en&btnG=Search&q=intitle:The+Genetic+Information+Nondiscrimination+Act+of+2008+and+the+Patient+Protection+and+Affordable+Care+Act+of+2010+:+Overview+and+Legal+Analysis+of+Potential+Interactions#2. Accessed 11 June 2014.
Etheredge LM. A rapid-learning health system. Health Aff (Millwood). 2007;26(2):w107–18. doi:10.1377/hlthaff.26.2.w107.
Mello MM, Wolf LE. The Havasupai Indian tribe case: lessons for research involving stored biological samples. N Engl J Med. 2010;363(3):204–7.
Sarda S, Hannenhalli S. Next-generation sequencing and epigenomics research: a hammer in search of nails. Genomics Inform. 2014;12(1):2–11. doi:10.5808/GI.2014.12.1.2.
Acknowledgments
AA Nishimura reports Grants from National Center for Advancing Translational Sciences, National Institutes of Health (funded under NCATS Grant TL1 TR000422). The focus of this paper overlaps with the work of NIH/NGRI U01 HG006507 which is one of the funding sources for P Tarczy-Hornoch and BH Shirts.
Disclosure
AA Nishimura and P Tarczy-Hornoch both declare no conflicts of interest. BH Shirts has received a consulting fee from Avalon Healthcare Solutions.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nishimura, A.A., Tarczy-Hornoch, P. & Shirts, B.H. Pragmatic and Ethical Challenges of Incorporating the Genome into the Electronic Health Record. Curr Genet Med Rep 2, 201–211 (2014). https://doi.org/10.1007/s40142-014-0051-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40142-014-0051-9