Abstract
Introduction
The aim was to assess the performance of a blood assay combining measurements of MxA (myxovirus resistance protein A) and CRP (C-reactive protein) to differentiate viral from bacterial respiratory infections.
Methods
In a prospective study, MxA and CRP were measured in the blood by the AFIAS panel in adults admitted with respiratory infection. Patients were split into discovery and validation cohorts. Final diagnosis was adjudicated by a panel of experts. Microbiology-confirmed cases comprised the discovery cohort, and infections adjudicated as highly probable viral or bacterial comprised the validation cohort.
Results
A total of 537 patients were analyzed: 136 patients were adjudicated with definitive viral infections and 131 patients with definitive bacterial infections. Using logistic regression analysis, an equation was developed to calculate the probability for bacterial infection using the absolute value of MxA and CRP. Calculated probability ≥ 0.5 and/or MxA to CRP ratio less than 2 applied as the diagnostic rule for bacterial infections. This rule provided 91.6% sensitivity and 90.4% negative predictive value for the diagnosis of bacterial infections. This diagnostic sensitivity was confirmed in the validation cohort. A MxA/CRP ratio less than 0.15 was associated with unfavorable outcome.
Conclusion
The calculation of the probability for bacterial infection using MxA and CRP may efficiently discriminate between viral and bacterial respiratory infections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Why carry out this study? |
There is an unmet need to differentiate if an acute respiratory infection is of bacterial or viral etiology. |
The existence of a rapid blood test would prevent unnecessary consumption of antibiotics. |
This study investigates if blood measurements of MxA (myxovirus resistance protein A) and CRP (C-reactive protein) can inform if a respiratory tract infection is caused by bacteria, viruses, or both. |
What was learned from the study? |
MxA and CRP can be used for the differential diagnosis between viral and bacterial respiratory tract infections. Quantitative blood results are interpreted using the calculated probability for bacterial infection and taking into consideration the ratio of MxA to CRP. |
The diagnostic algorithm needs to be evaluated into a randomized controlled trial to guide prescription of antibiotics. |
Introduction
Viral and bacterial infections represent a major source of morbidity, mortality, and healthcare costs. Approximately 80% of all antibiotics are prescribed at a primary healthcare setting and the majority are for respiratory tract infections [1]. Most acute respiratory infections are of viral etiology and a tool is required to guide clinicians to avoid unnecessary antibiotic prescription. This tool should be point-of-care (POC) and readily distinguish between viral and bacterial infections.
Procalcitonin (PCT) is the most broadly used biomarker to guide appropriate prescription of antibiotics for respiratory infections [2]. The evaluation of the patient several times looks like a scale where one edge represents the likelihood to have viral infection and the other edge to have bacterial infection. Each edge needs to be represented by biomarkers, one of which should indicate the likelihood for viral infection and another the likelihood for bacterial infection. This need is dictated by several microbiology studies showing that infections of the lower respiratory tract are often of mixed viral and bacterial etiology [3, 4].
Candidate biomarkers are myxovirus resistance protein A (MxA) and C-reactive protein (CRP). Mx proteins are large interferon-induced GTPases involved in the control of intracellular pathogens [5]. In humans, two Mx homologs mediate antiviral activity against a broad range of viruses, including SARS-CoV-2 [6]. Elevated MxA indicates increased endogenous interferon production mediated by viral activation, and it can be used as a marker of viral infection [6]. CRP is a well-described cytokine-induced acute-phase protein; blood levels increase following non-specific responses to infections stimuli [1]. These two protein measurements are integrated into one simply POC device which informs in a qualitative approach if MxA, CRP, or both are increased [7].
The limitation of qualitative approaches is that they neglect the individualized nature of patients, several of whom are often co-infected by viruses and bacteria. The present study followed a novel design to investigate the value of MxA and CRP as diagnostic tools of the etiology of respiratory infections. Continuous measurements of both proteins were analyzed and patients were split into a discovery cohort and a validation cohort. The discovery cohort was composed of patients with definitive infections and the validation cohort of patients with high probability for infections of viral etiology or bacterial etiology or for co-infection. Cut-offs developed in the discovery cohort were validated in the validation cohort.
Patients and methods
This is a prospective study which was conducted between July 2022 and February 2023 in six departments of internal medicine located at public hospitals in Greece. The study was a sub-study part of the clinical trials ACCESS and ImmunoSep, which were licensed by the National Ethics Committee of Greece (approvals 122/20 and 2/21) and the National Organization for Medicines of Greece (approvals IS113/20 and IS008/21) (ClinicalTrials.gov registrations NCT04724044 and NCT04990232). Written informed consent was provided by the patients or their legal representative. Patients who were screened for eligibility for both trials participated in the present study. The study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments.
Enrolled patients should meet both the following inclusion criteria: (1) male or female adults (age 18 years or more), and (2) any of the following clinical signs of infection: fever, cough, dyspnea, sore throat, headache, nasal congestion, or diarrhea. Exclusion criteria were: age less than 18 years; chronic oral or intravenous intake of corticosteroids at a dose more than 0.4 mg/kg of equivalent prednisone daily; known infection by the human immunodeficiency virus; neutropenia; any chronic anti-cytokine treatment; and pregnancy or lactation.
Forty patients who did not meet any of the exclusion criteria and who did not present any sign of active infection were enrolled as comparators.
All the study participants were subject to an intense work-out, including present and past medical history and thorough physical examination; laboratory evaluation including complete blood cell counting, biochemistry, blood gas and procalcitonin; chest X-ray, and, when required, high-resolution chest computed tomography; nasopharyngeal swab collection for real-time PCR testing for SARS-CoV-2 and, when needed, by the BioFire® FilmArray® panel of the upper or lower respiratory tract (Respiratory Panel 2.1 plus or Pneumonia plus Panel respectively) (bioMérieux, Lyon France); urine detection of the antigens of Streptococcus pneumoniae and Legionella by the BinaxNOW assay (Abbott Point of care); glutamate dehydrogenase and toxin A/B immunosorbent assays for Clostridioides difficile in the stool; and blood, sputum, or urine culture. The SOFA (sequential organ failure assessment) score and the Charlson’s comorbidity index (CCI) were calculated. Patients were followed-up for 28 days for survival.
In parallel to routine blood sampling, another 4 mL of venous whole blood was collected into one EDTA-coated tube (Vacutainer; Becton Dickinson, Cockeysville MD, USA) and analyzed using the AFIAS MxA/CRP test.
AFIAS MxA/CRP is a fluorescent lateral flow immunoassay test for the quantitative determination of MxA and CRP in human blood, consisting of an all-in-one cartridge with a detector part, a diluent part, and the test strip with anti-human MxA, anti-human CRP, and CRP antigen in the test line and chicken IgY in the control line. The test was run on an AFIAS-10 automated immunoassay analyzer by Boditech Med (Chuncheon, Republic of Korea). The lower limit of detection was 10 ng/mL for MxA and 1 mg/L for CRP.
Patients were classified into seven categories following adjudication by two experts, who were given access to all clinical and laboratory information for the patients 30 days after completion of the follow-up. The only non-accessible information was the result of AFIAS MxA/CRP. Adjudicators were told to classify patients into one the following seven categories using their clinical judgment and available laboratory and microbiology results. In case the two experts did not agree, a third expert served as an arbiter: (1) no infection: patients without infection; (2) definitive viral infection: infection definitively caused by an isolated virus and where no bacterial pathogen was detected; (3) definitive bacterial infection: infection definitively caused by an isolated bacterial species and where no viral pathogen was detected; (4) definitive viral/bacterial co-infection: infection definitively caused by both viral and bacterial species; (5) high probability for viral infection: infection most probably of viral etiology without any viral pathogen detected, (6) high probability of bacterial infection: infection most probably of bacterial etiology without any bacterial pathogen detected, and (7) definitive viral infection and high probability for bacterial co-infection: infection definitively caused by an isolated virus with high probability of bacterial co-infection without, however, any bacterial pathogen detected.
The study primary endpoint was to develop an algorithm which uses blood levels of MxA, CRP, and their ratio to classify patients into viral and bacterial infections or co-infection.
The study's secondary endpoint was to develop cut-offs of MxA, CRP, and their ratio for the early prognosis of a 28-day unfavorable outcome.
Qualitative variables were expressed as frequencies and 95% confidence intervals (CIs) and qualitative variables as medians and 95% CIs. Comparisons of qualitative variables between groups were carried out by Fisher’s exact test. Comparisons of quantitative variables between groups were carried out by the Mann–Whitney U test after Bonferroni correction for multiple testing. Patients with definitive viral and bacterial infections were the discovery cohort. In these patients, two analyses were carried out: (1) the diagnostic performance of the combination of the suggested diagnostic cut-offs for MxA (> 15 ng/mL) and for CRP (> 10 mg/L) were determined; and (2) logistic regression analysis was performed to provide an equation which can define the probability of a patient for bacterial or viral infection. Then, a receiver operator characteristic (ROC) curve analysis was carried out to define a cut-off of the probability with the best trade-off using the Youden index. The area under the curve (AUC) and the 95% CIs were calculated. In parallel, the ratio of MxA to CRP was calculated and plotted as the ROC curve, which provides the best trade-off for the diagnosis of viral infection. Then, the two cut-offs, the cut-off of probability for bacterial infection and the cut-off of the MxA/CRP ratio, were combined to a final diagnostic rule for the discrimination between bacterial and viral infections. This rule was validated at the validation cohort. Finally, these cut-offs were applied for patients with definitive viral/bacterial co-infection and for patients with definitive viral and highly probable bacterial co-infection. Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated. Any p value below 0.05 was considered statistically significant.
Results
A total of 537 patients were enrolled (Table 1), 40 of whom without infection were used as comparators. A total of 267 patients were included at the discovery cohort: 136 had definitive viral infection and 131 had definitive bacterial infection (Fig. 1). The remaining patients were analyzed at the validation cohort. Overall, patient mean age was 68.1 years, and 60.7% were men. The mean SOFA was 3.27 and the mean CCI 3.9. Overall, 28-day mortality was 19.0%.
The diagnostic performance of MxA and CRP was analyzed in the total of enrolled patients using the cut-off values suggested by the manufacturer. MxA levels ≥ 15 ng/mL could discriminate viral infection with 79.7% sensitivity and 80.0% specificity versus no infection (Fig. 2A). CRP ≥ 10 mg/L had 96.7% sensitivity and 65.0% specificity for the definitive bacterial infection group versus no infection (Fig. 2B).
Diagnostic performance of MxA and CRP based on pre-defined cut-offs for bacterial and viral infections. A Diagnostic performance of the MxA in definitive viral infection group; definitive viral infection (n = 136), definitive viral/bacterial co-infection (n = 20), definitive viral/high probability bacterial co-infection (n = 71). B Diagnostic performance of the CRP in definitive bacterial infection group; definitive bacterial infection (n = 131), definitive viral/bacterial co-infection (n = 20). The confidence intervals (95% CIs) of each percentage are provided in parentheses. CRP C-reactive protein, MxA myxovirus resistance protein A, n number of patients, NPV negative predictive value, PPV positive predictive value
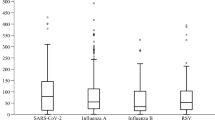
The primary endpoint was first analyzed in the discovery cohort of 267 patients with definitive adjudication as either viral infection or bacterial infection. Concentrations of MxA were greater in viral infections and of CRP in bacterial infections (Fig. 3A). Following logistic regression analysis, an equation was generated which could take into consideration the absolute values of MxA and CRP and conclude with the probability for bacterial infection (Supplementary Figs. 1A to 1C). The ROC of this probability could clearly separate bacterial from viral infections when values were 0.5 or more (Supplementary Fig. 2). To further strengthen the diagnostic performance of this probability, the ROC curve of the MxA/CRP ratio was calculated, and values of 2 or more could separate viral from bacterial infections (Supplementary Fig. 3). Then, we used the probability and MxA/CRP in one single diagnostic rule, according to which patients with probability 0.5 or more and/or MxA/CRP less than 2 had bacterial infection. This rule provided sensitivity of 91.6% and NPV 90.4% for bacterial infection (Fig. 3B). At this stage, we thought that the NPV greater than 90% was important since it might prevent missing patients in need of antibiotics.
MxA, CRP, and MxA/CRP ratio for the differential diagnosis between definitive bacterial and viral infection at the discovery cohort. A Blood concentrations of MxA and CRP of comparators without infection (n = 40), of patients with definitive viral infection (n = 136), and of patients with definitive bacterial infection (n = 131). Circles denote extreme values and asterisks denote outliers #p < 0.001 versus no infection; ##p < 0.0001 versus definitive viral infection. B Diagnostic performance of the combination of MxA and of CRP using the rule which combines the probability for bacterial infection (P) and the MxA/CRP ratio. Analysis involved the total of 267 patients with either definitive viral infection or definitive bacterial infection. The confidence intervals of each percentage are provided in parentheses. CRP C-reactive protein, n number of patients, MxA myxovirus resistance protein A, NPV negative predictive value, PPV positive predictive value, Se sensitivity, Sp specificity
In the validation cohort, MxA was greater among patients with high probability for viral infections and CRP was greater among patients with high probability for bacterial infections (Fig. 4). When the cut-offs developed at the discovery cohort were applied, the calculated sensitivity for bacterial infections was 89.1% and the PPV for bacterial infections was 97.4%. However, the NPV for bacterial infections was lower than the discovery cohort (Table 2).
MxA and CRP for the differential diagnosis between high probability bacterial infection and high probability viral infection in the validation cohort. Blood concentrations of MxA and CRP of patients with high probability for viral infection (n = 11) and of patients with high probability for bacterial infection (n = 128) are provided. Circles denote extreme values and asterisks denote outliers
#p < 0.05 versus high probability for bacterial infection; ##p < 0.0001 versus high probability for viral infection. CRP C-reactive protein, n number of patients, MxA myxovirus resistance protein A
Both MxA and CRP were high among patients with co-infections (Fig. 5). It was found that, among the 71 patients with definitive viral infection and high probability for bacterial co-infection, 46 (64.8%) were positive for the diagnostic rule which is using the calculated probability and the MxA/CRP ratio. This rule was also positive in 14 of the 20 patients (70%) with definitive viral/bacterial co-infection.
MxA and CRP for the diagnosis of viral/bacterial co-infection. Blood concentrations of MxA and CRP of patients with definitive viral infection and high-probability for bacterial infection (n = 71) and of patients with definitive viral/bacterial co-infection (n = 20). #p: 0.351 versus high probability for bacterial infection; ##p: 0.413 versus high probability for viral infection. CRP C-reactive protein, n number of patients, MxA myxovirus resistance protein A
MxA was lower in non-survivors than in survivors (Fig. 6A) and CRP was higher in non-survivors (Fig. 6B). The AUC of the ROC curve was higher for the MxA/CRP ratio than for MxA for the prediction of unfavorable outcome after 28 days (Fig. 6C). Values lower than 0.15 were associated with NPV 85.4% for the exclusion of risk for death (Fig. 6D).
MxA/CRP ratio for the prognosis of unfavorable outcome. A Comparison of MxA between 28-day survivors and 28-day non-survivors; the p value of comparison is provided. B Comparison of CRP between 28-day survivors and 28-day non-survivors; the p value of comparison is provided. C ROC curves of MxA and MxA/CRP ratio for the prognosis of unfavorable outcome. D Prognostic performance of the MxA/CRP ratio less than 0.15 for death after 28 days; the OR and 95% confidence intervals for death with MxA/CRP less than 0.15 are provided. Analysis involves the total of 537 studied patients. AUC area under the curve, CI confidence interval, CRP C-reactive protein, MxA myxovirus resistance protein A, n number of patients, NPV negative predictive value, OR odds ratio, PPV positive predictive value, ROC receiver operating characteristics
Discussion
The present study provides evidence that the combination of MxA and CRP under a specific algorithm may safely discriminate viral from bacterial infections. The developed algorithm contains the integration of calculated probability and of the ratio MxA to CRP. In this approach, the overall sensitivity and the overall NPV for bacterial infections go beyond 90%.
This is not the first study in which MxA and CRP have been used in parallel for the differential diagnosis between viral and bacterial infections. Published evidence is mainly coming from one POC device which is using blood from the fingertip applied on a cartridge. The cartridge is running an immunoassay and the results are analyzed by a reader providing qualitative interpretation. Available publications describe lower numbers of patients than the present study. The first study enrolled patients with acute respiratory symptoms: 34 with COVID-19; 1 infected by a virus other than SARS-CoV-2; 8 with bacterial pneumonia; and 4 with non-infectious lung disorders. The reported sensitivity for the detection of viral infection was 100% and of bacterial infection 100%. However, the study enrolled limited number of patients [8]. In a study of only 54 patients, the sensitivity for the diagnosis of bacterial respiratory infection was 80% [9].
The largest study on the utility of MxA and CRP as tools to discriminate between viral and bacterial acute respiratory illness in the emergency department was presented for 520 patients. Analysis focused most on the sensitivity and specificity for the diagnosis of bacterial infections which was reported as 93.2% and 88.4%, respectively [10]. This diagnostic performance is in accordance with our findings. The authors suggest that the use of this tool may reduce the consumption of antibiotics from 18.4% to 11.6%.
One recent meta-analysis described 421 articles on the use of POC tools for diagnostic purposes in the emergency department. The results showed that single biomarkers have sub-optimal sensitivity for bacterial infections. However, studies on the combination of MxA and CRP were not included in the meta-analysis [11]. The results of this meta-analysis strengthen the need for the application of at least two biomarkers. We believe that the balance between the biomarkers should be taken into consideration, and this is expressed by the MxA/CRP ratio in our study. Furthermore, the results provided to the clinician staff should not only be limited to the qualitative interpretation for MxA and CRP but quantitative data are needed. Indeed, in a recent study of 200 patients, quantitative blood measurements of MxA and CRP were matched to qualitative interpretation (low or high). Although most patients reported as low MxA or low CRP had low absolute values, some patients were false positive or false negative, i.e., they were reported as low MxA/CRP but the absolute values were increased and vice versa [12]. We strongly suggest that patients should be subjected to quantitative measurements followed by an artificial intelligence interpretation algorithm.
One limitation of the present study is the inclusion of patients infected by SARS-CoV-2. It is well known that COVID-19 infection increases the production of CRP either by the virus per se or by bacterial co-infection [13, 14]. Further studies are required to further clarify if MxA and CRP may assist in the detection of bacterial co-infection in patients with COVID-19.
Conclusion
The present study presents evidence that the combination of MxA and CRP may assist in the differential diagnosis between viral and bacterial respiratory infections. Interpretation of the measurements should rely on the suggested algorithm which uses the calculated probability and the ratio of MxA to CRP. This algorithm needs to be evaluated into a randomized controlled trial to guide prescription of antibiotics.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Joseph P, Godofsky E. Outpatient antibiotic stewardship: a growing frontier-combining myxovirus resistance protein A with other biomarkers to improve antibiotic use. Open Forum Infect Dis. 2018;5:ofy024.
Kyriazopoulou E, Poulakou G, Giamarellos-Bourboulis EJ. Biomarkers in sepsis: can they help improve patient outcome? Curr Opin Infect Dis. 2021;34:126–34.
Jain S, Self WH, Wunderink RG, et al. Community-acquired pneumonia requiring hospitalization among US adults. N Engl J Med. 2015;373:415–27.
Kyriazopoulou E, Karageorgos A, Liaskou-Antoniou L, et al. BioFire®FilmArray® pneumonia panel for severe lower respiratory tract infections: subgroup analysis of a randomized clinical trial. Infect Dis Ther. 2021;10:1437–49.
Gao S, von der Malsburg A, Dick A, et al. Structure of myxovirus resistance protein A reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity. 2011;35:514–25.
Dick A, Graf L, Olal D, et al. Role of nucleotide binding and GTPase domain dimerization in dynamin-like myxovirus resistance protein A for GTPase activation and antiviral activity. J Biol Chem. 2015;290:1277992.
Shirley M. FebriDx®: a rapid diagnostic test for differentiating bacterial and viral aetiologies in acute respiratory infections. Mol Diagn Ther. 2019;23:803–9.
Karim N, Ashraf MZ, Naeem M, et al. Utility of the FebriDx point-of-cate test for rapid triage and identification of possible coronavirus disease 2019 (COVID-19). Int J Clin Pract. 2020;75: e13702.
Sambursky R, Shapiro N. Evaluation of a comined MxA and CRP point-of-care immunoassay to identify viral and/or bacterial immune response in patients with acute febrile respiratory infection. Eur Clin Resp J. 2015;2:28245.
Shapiro NI, Filbin MR, Hou PC, et al. Diagnostic accuracy of bacterial and viral biomarker point-of-care test in the outpatients setting. JAMA Netw Open. 2022;5: e2234588.
Gentilotti E, De Nardo P, Cremonini E, et al. Diagnostic accuracy of point-of-care tests in acute community-acquired lower respiratory tract infections. A systematic review and meta-analysis. Clin Microbiol Infect. 2022;28:13–22.
Akelew Y, Derbew A, Lemma M, et al. Evaluation of C-reactive protein and myxovirus resistance protein A to guide the rational use of antibiotics among acute febrile adult patients in Northwest Ethiopia. Int J Infect Dis. 2020;101:276–82.
Rizzi M, D’Onghia D, Tonello S, et al. COVID-19 biomarkers at the crossroad between patient stratification and targeted therapy: the role of validated and proposed parameters. Int J Mol Sci. 2023;24:7099.
Kim JYH, Ragusa M, Tortosa F, et al. Viral reactivations and co-infections in COVID-19 patients: a systematic review. BMC Infect Dis. 2023;23:259.
Funding
The study was funded in part by the Hellenic Institute for the Study of Sepsis and by Boditech Inc, South Korea.
Author information
Authors and Affiliations
Contributions
Conceptualization: Evangelos J Giamrellos-Bourboulis. Methodology: Konstantina Iliopoulou, Panagiotis Koufargyris, Sarantia Doulou, Elisavet Tasouli, Sokratis Katopodis, Stavroula-Porphyria Chachali, Georgios Schinas, Charalambos Karachalios, Myrto Astriti, Parakevi Katsaounou, George Chrysos, Theodors Seferlis, Effrosyni Dimopoulou, Myrto Kollia, Garyphalia Poulakou, Styliani Gerakari, Ilias C Papanikolaou, Haralampos Milionis, George N Dalekos, Vasiliki Tzavara, Theano Kontopoulou. Formal analysis and investigation: Evangelos J Giamrellos-Bourboulis. Writing—original draft preparation: Evangelos J Giamrellos-Bourboulis. Writing—review and editing: Konstantina Iliopoulou, Panagiotis Koufargyris, Sarantia Doulou, Elisavet Tasouli, Sokratis Katopodis, Stavroula-Porphyria Chachali, Georgios Schinas, Charalambos Karachalios, Myrto Astriti, Parakevi Katsaounou, George Chrysos, Theodors Seferlis, Effrosyni Dimopoulou, Myrto Kollia, Garyphalia Poulakou, Styliani Gerakari, Ilias C Papanikolaou, Haralampos Milionis, George N Dalekos, Vasiliki Tzavara, Theano Kontopoulou. Funding acquisition: Evangelos J Giamrellos-Bourboulis Resources: Evangelos J Giamrellos-Bourboulis. Supervision: Evangelos J Giamrellos-Bourboulis.
Corresponding author
Ethics declarations
Conflict of interest
Garyphalia. Poulakou has received honoraria and/or consulting fees by Astra-Zeneca, Gilead, GSK, Menarini, MSD, Norma, Pfizer and SOBI and research grants by the University of Minnesota/University College London, the Hellenic Institute for the Study of Sepsis, Bausch, Roche, Xenothera, FabNTech and Pfizer. Ilias C. Papanikolaou has received honoraria or served as PI for studies from Boehringer-Ingelheim, GlaxoSmithKline and AstraZeneca. Haralampos. Milionis reports receiving honoraria, consulting fees and non-financial support from healthcare companies, including Amgen, Angelini, Bayer, Mylan, MSD, Pfizer, and Servier. George N Dalekos is an advisor or lecturer for Pfizer, Roche, Sanofi and Sobi, and received research grants from Gilead and has served as PI in studies for Gilead, Novo Nordisk, Genkyotex, Regulus Therapeutics Inc, Tiziana Life Sciences, Bayer, Astellas, Pfizer, Amyndas Pharmaceuticals, CymaBay Therapeutics Inc., Sobi and Intercept Pharmaceuticals. Evangelos. J. Giamarellos-Bourboulis has received honoraria from Abbott Products Operations, bioMérieux, Brahms GmbH, GSK, InflaRx GmbH, Sobi and Xbiotech Inc; independent educational grants from Abbott Products Operations, bioMérieux Inc, InflaRx GmbH, Johnson & Johnson, MSD, Sobi and Xbiotech Inc.; and funding from the Horizon2020 Marie Skłodowska-Curie International Training Network “the European Sepsis Academy” (granted to the National and Kapodistrian University of Athens), the Horizon 2020 European Grants ImmunoSep and RISCinCOVID and the Horizon Health grant EPIC-CROWN-2 (granted to the Hellenic Institute for the Study of Sepsis). Konstantina Iliopoulou, Panagiotis Koufargyris, Sarantia Doulou, Elisavet Tasouli, Sokratis Katopodis, Stavroula-Porphyria Chachali, Georgios Schinas, Charalambos Karachalios, Myrto Astriti, Parakevi Katsaounou, George Chrysos, Theodors Seferlis, Effrosyni Dimopoulou, Myrto Kollia, Styliani Gerakari, Vasiliki Tzavara, and Theano Kontopoulou have no competing interests.
Ethical approval
The study is a sub-study part of the clinical trials ACCESS and ImmunoSep which were licensed by the National Ethics Committee of Greece (approvals 122/20 and 2/21) and the National Organization for Medicines of Greece (approvals IS113/20 and IS008/21) (ClinicalTrials.gov registrations NCT04724044 and NCT04990232). Written informed consent was provided by the patients or their legal representative. Patients who were screened for eligibility for both trials participated in the present study. The study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Iliopoulou, K., Koufargyris, P., Doulou, S. et al. Developing a Tool for Differentiation Between Bacterial and Viral Respiratory Infections Using Myxovirus Resistance Protein A and C-Reactive Protein. Infect Dis Ther 13, 105–119 (2024). https://doi.org/10.1007/s40121-023-00901-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40121-023-00901-2