Abstract
Epidemic IncF plasmids have been pivotal in the selective advantage of multidrug-resistant (MDR) extraintestinal pathogenic Escherichia coli (ExPEC). These plasmids have offered several advantages to their hosts that allowed them to coevolve with the bacterial host genomes and played an integral role in the success of ExPEC. IncF plasmids are large, mosaic, and often contain various types of antimicrobial resistance (AMR) and virulence associated factor (VAF) genes. The presence of AMR, VAF genes, several addition/restriction systems combined with truncated transfer regions, led to the fixation of IncF plasmids in certain ExPEC MDR clones, such as ST131 and ST410. IncF plasmids entered the ST131 ancestral lineage in the mid 1900s and different ST131 clade/CTX-M plasmid combinations coevolved over time. The IncF_CTX-M-15/ST131-C2 subclade combination emerged during the early 2000s, spread rapidly across the globe, and is one of the greatest clone/plasmid successes of the millennium. The ST410-B3 subclade containing blaCTX-M-15 incorporated the NDM-5 carbapenemase gene into existing IncF platforms, providing an additional positive selective advantage that included the carbapenems. A “plasmid-replacement” clade scenario occurred in the histories of ST131 and ST410 as different subclades gained different AMR genes on different IncF platforms. The use of antimicrobial agents will generate selection pressures that enhance the risks for the continuous emergence of MDR ExPEC clone/IncF plasmid combinations. The reasons for clade/IncF replacements and associations between certain clades and specific IncF plasmid types are unknown. Such information will aid in designing management and prevention strategies to combat AMR.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Epidemic IncF plasmids have been pivotal in the selective advantage of certain extraintestinal pathogenic Escherichia coli (ExPEC) clones such as ST131 and ST410. |
IncF plasmids are large, mosaic, and often contain various types of antimicrobial resistance (AMR) and virulence associated factor (VAF) genes. |
The presence of AMR, VAF genes, several addition/restriction systems, combined with truncated transfer regions, has led to fixation of IncF plasmids in certain ExPEC multidrug (MDR) clones, such as ST131 and ST410. |
IncF plasmids entered the ST131 ancestral lineage in the mid 1900s and different ST131 clade/CTX-M plasmid combinations coevolved over time. The IncF_CTX-M-15/ST131-C2 subclade combination emerged during the early 2000s, spread rapidly across the globe, and is one of the greatest clone/plasmid successes of the millennium. |
The ST410-B3 subclade containing blaCTX-M-15 incorporated the NDM-5 carbapenemase gene into existing IncF platforms, providing an additional positive selective advantage that included the carbapenems. |
The use of antimicrobial agents likely generates selection pressures that enhance the risks for the continuous emergence of successful MDR ExPEC clone/IncF plasmid combinations. |
Introduction
Antimicrobial resistance (AMR) is a substantial peril to human health [1]. Without effective antibiotics, modern medicine will cease to function in its current state, as chemotherapy and routine surgery will be deemed too high risk for the fear of non-treatable infections due to multidrug-resistant (MDR) bacteria. In 2019, an estimated 4.95 million deaths worldwide were associated with bacterial AMR, including 1.27 million deaths attributable to bacterial AMR [2]. There is an urgent need to preserve the world’s remaining antibiotics, especially effective and cost-effective drugs.
The persistence of AMR genes within bacterial populations is due to the perseverance of certain successful MDR clones and/or the movement of AMR genes within and between diverse strains or clones [3, 4]. Bacterial MDR high-risk clones act as “hoarders and spreaders” of AMR genes through horizontal and vertical transmission [3]. The capture and intracellular movement of AMR genes occurs via mobile genetic elements (MGEs) such as insertion sequence elements, transposons, and integrons [5]. The intercellular movement of AMR genes is due to different types of MGEs that include plasmids, integrative conjugative elements, and bacteriophages [5].
Horizontal gene transfer has had a major impact on bacterial evolution and diversification by allowing gene flow between different isolates of the same species or between various species [6, 7]. Plasmids are important vehicles for horizontal gene transfer [8]. They usually move genes between bacterial cells through conjugation [7, 8]. This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Plasmid Basics
Plasmids are double-stranded, extrachromosomal DNA, mostly circular, self-replicating genetic elements in Gram-positive and Gram-negative bacteria that often provide accessory genes to the host bacteria [8]. They differ in sizes, copy number, replication mechanisms, transmission modes, and host range [7]. Plasmids can broadly be classified into two groups, namely plasmids with low copy numbers and plasmids with high copy numbers [7]. Low copy number plasmids are frequently large in size (i.e., > 50 kb), conjugative (i.e., ability to move between bacterial isolates), and within their hosts they are detected in low numbers (i.e., < 10 copies per host) [7]. High copy number plasmids, in general, are smaller (i.e., < 40 kb), typically lack functional conjugative systems (i.e., do not have the ability to move between bacterial cells by their own means), and within their hosts they are detected in high numbers (i.e., > 20 copies per host). Other classification systems that have been published include those comparing nucleotide homologies, replication systems, and mobility proteins of plasmids [7].
Plasmids contain genes coding for features that promote their own survival [7]. These genes are responsible for the maintenance of plasmids and are referred to as “core” or “backbone” genes. Some examples include genes coding for replicative functions, conjugative functions, anti-restriction proteins, and post-segregational killing systems. Plasmids can also contain other types of genes, referred to as accessory genes, that provide ecologically selective advantages to their hosts [9]. Accessory genes include AMR, heavy metal resistance, metabolic and virulence associated factor (VAF) genes. The spread of accessory genes via plasmids has been pivotal in the evolution and survival of different bacterial species, especially among members of the Enterobacterales [7, 9].
Conjugative plasmids contain genes coding for their own means of transmission [5]. They rely heavily on the host’s machinery for their successful replication and conjugation and are a fitness burden to the host [9]. The structure of conjugative plasmids is broadly divided into four regions [5]: (i) The ori (origin of replication) that is responsible for the replication of plasmids. (ii) The oriT (origin of transfer) and genes encoding relaxase, type IV coupling protein (T4CP), and type IV secretion system (T4SS). These structures are responsible for the conjugation of plasmids between bacterial isolates. (iii) Regions responsible for anti-restriction proteins and post-segregational killing systems. (iv) Accessory genes that often consist of AMR, VAF genes and their associated flanking regions.
Several plasmid typing schemes are available and two are often used to classify plasmids [10]. The first scheme is named “replicon or rep” typing that targets the ori (i.e., responsible for plasmid replication). “Rep” typing divides plasmids into groups (e.g., RepA, RepC, etc.). Plasmids with the same replicon cannot coexist within the same host, hence the name “incompatibility” or “Inc” groups (e.g., IncA, IncC etc.). “Inc” terminology is more often used in published literature than Rep groups [10]. The second common typing scheme is “plasmid multilocus sequencing” (pMLST) that targets specific conserved structures within certain Inc plasmids [11]. pMLST is often used to subtype certain Inc plasmids such as IncC, IncI1, IncHI1, IncHI2, IncF, and IncN. For conjugative plasmids, two additional schemes, mobility (MOB) and mating pair formation (MPF) typing, based on the relaxase and T4SS virB4 amino acid sequences, were also developed, but they were less frequently used in the literature than Rep typing [12,13,14].
Some Inc plasmid types are distributed across various bacterial species and among different clones of the same species and are referred to as “broad host range” plasmids (e.g., IncP and IncO) [15]. Broad host range plasmids contain efficient conjugation systems enabling such plasmids to move efficiently between different bacterial species and be stably maintained [16]. “Narrow-host range” plasmids (e.g., IncF) can only be transferred between and maintained in closely related species or clones [15]. The current understanding of the plasmid host ranges has primarily been based on the studies from certain “model” plasmids and the specific host ranges of clinically important AMR plasmids remain to determined.
The term “epidemic” refers to plasmids that are highly prevalent among various bacterial species obtained from different countries, over prolonged time periods, and from different One Health settings (e.g., human, animals, environment) [4]. “Epidemic plasmids” can either be high or low copy number, have broad or narrow host range distribution, or be conjugative or non-conjugative. It is important to keep in mind that standard definitions of what constitute “epidemic plasmids” (i.e., number of different bacterial species, prevalence [percentage], geographical distribution [how many countries], and prolonged time periods [months or years]) are currently lacking [4]. This needs to be addressed by an international expert committee.
The Plasmid Paradox
Plasmids are found frequently and widely among bacteria, even in the absence of positive selection pressure for traits/advantages provided by the plasmid’s accessory genes. This is known as the “plasmid paradox” [17]. Plasmids are metabolic burdens within their hosts that lead to fitness costs. Such fitness costs, combined with flawed plasmid segregation, suggest that plasmids will eventually be lost from bacterial populations over time. In the absence of sufficient positive selection pressures, daughter cells without plasmids should be able to eventually outcompete daughter cells containing plasmids [17]. If the plasmid-encoded accessory traits are beneficial for the continuing survival of the hosts (i.e., the positive selection pressures are strong enough to overcome the plasmid fitness costs) such beneficial accessory genes will eventually be captured and incorporated into the bacterial chromosome over time [17]. Therefore, in theory at least, plasmids should not be able to survive for long periods of time within bacterial hosts, even in the presence of constant positive selection pressure. Brockhurst and Harrison recently provided several ecological and evolutionary solutions to the plasmid paradox [18]. These included infectious transmission, variation in fitness costs, compensatory evolution in hosts and on plasmids, and “piggybacking” niche adaptions. The current challenge for microbiologists as well as population and evolutionary biologists is to determine how these factors contribute to the successful survival of plasmids within bacterial populations, over prolonged periods of time.
Antimicrobial Resistance Plasmids
The horizontal spread of AMR genes between bacteria is mainly due to conjugative plasmids [8]. Historically, conjugative plasmids containing AMR genes were referred to as R plasmids [19]. The clinical importance of AMR was a major driving force in the genomic analysis of R plasmids and was responsible for establishing current plasmid biology foundations.
The positive selective benefits of R plasmids are readily apparent. AMR genes offer immediate advantage to their respective hosts when exposed to antimicrobial pressures [9]. Some large R plasmids contain various AMR genes relating to different antibiotic classes. This is evident among the Enterobacterales where different AMR genes, on the same large plasmids, are responsible for resistance to the following different antibiotic classes: β-lactam antibiotics (e.g., β-lactamases such as blaTEM), aminoglycosides (e.g., aminoglycoside-modifying enzymes such as aad), fluoroquinolones (e.g., plasmid-mediated quinolone resistance determinants such as qnr), tetracyclines (e.g., tet genes), and trimethoprim-sulfamethoxazole (e.g., sul and dfr genes) [8]. The clinical use of unrelated agents (i.e., β-lactams and tetracyclines) created sufficient positive selective advantages for maintenance of different AMR genes within bacterial populations.
Extraintestinal Pathogenic Escherichia coli (ExPEC) Pathotypes
E. coli is divided into three pathotypes [20], namely (i) Commensal flora, i.e., isolates that are normal flora of the gastrointestinal tract (GIT) of humans and animals. (ii) Diarrheagenic E. coli, i.e., isolates that cause GIT infections. (iii) Extraintestinal pathogenic E. coli (ExPEC), i.e., Isolates that cause infections outside the GIT. The ExPEC pathotype consists of the following sub-pathotypes [20]: (a) Uropathogenic E. coli that are mainly responsible for urinary tract infections (UTIs). (b) Neonatal meningitis E. coli that are mainly responsible for central nervous system infections among neonates. (c) Sepsis-associated pathogenic E. coli that are linked with bloodstream infections. (d) Avian-pathogenic E. coli that causes infections in birds.
ExPEC have certain VAFs that differentiate this pathotype from commensal and diarrheagenic E. coli [20]. However, it seems these VAFs are not true virulence factors per se (as seen with diarrheagenic E. coli) but are more likely to be important in enhanced ecological survival, fitness, and play an important part in the evolution of ExPEC [20].
Human ExPEC is the most common cause of community-acquired and community-onset healthcare-associated urinary and bloodstream infections worldwide [21]. Thus, ExPEC evades the standard infection control and prevention practices that are used to contain hospital-acquired infections [22]. This E. coli pathotype is also a leading cause of sepsis, hospitalization, and death across the world, especially among the elderly [23]. Bloodstream infections caused by AMR ExPEC (especially isolates resistance to the fluoroquinolones and cephalosporins) are associated with increased mortality. AMR E. coli in 2019 was responsible for 850,000 global deaths [2]. AMR ExPEC isolates are also responsible for prolonged length of hospital stay with the subsequent increase in healthcare costs [24]. E. coli is also an important One Health (i.e., human, animal, environment) reservoir for AMR genes and is used to monitor the spread of such genes across One Health environments [25].
Multidrug-Resistant (MDR) High-Risk ExPEC Clones
In the 1990s, ExPEC obtained from human clinical infections was sensitive to most antibiotic classes [26]. During the 2000s, antimicrobial resistance increased exponentially, especially to fluoroquinolones (e.g., ciprofloxacin, norfloxacin, levofloxacin, moxifloxacin) and third-generation cephalosporins (i.e., cefotaxime, ceftriaxone, ceftazidime) [27]. The fluoroquinolones and third-generation cephalosporins are often used to treat serious ExPEC infections [28].
Mutations within the quinolone resistance determining regions (QRDR) confer resistance to the fluoroquinolones among ExPEC [29]. Three specific QRDR mutations are essential for high-level clinical resistance to the fluoroquinolones, namely gyrAS83L, gyrAD87N, and parCS80I. Plasmid-mediated fluoroquinolone resistance determinants (e.g., qnr, aac(6')-Ib-cr) also contributed to resistance of ExPEC to fluoroquinolones [29]. The most common causes of resistance to the third-generation cephalosporins among ExPEC are the extended-spectrum β-lactamases (ESBLs), more specifically the CTX-M enzymes [30]. The prevalence of CTX-M β-lactamases (especially CTX-M-15) has increased rapidly during the mid to late 2000s and are currently the most common global ESBL among E. coli and other members of the Enterobacterales [31]. The carbapenemases are the most important causes of carbapenem resistance because carbapenemase genes can be transferred between different Enterobacterales species through conjugative plasmids [32]. The most common carbapenemases among E. coli are the New Delhi metallo-β-lactamases (NDMs), OXA-48-like carbapenemase (especially OXA-48 and OXA-181), and Klebsiella pneumoniae carbapenemase (KPC) enzymes [33]. The VIM and IMP carbapenemases tend to be rare.
Certain ExPEC clones are overrepresented among non-selected E. coli populations (i.e., ST69, ST73, ST95, ST131), while other clones are overrepresented among MDR populations (i.e., ST131, ST405, ST38, ST648, ST410, ST167, and ST1193 [33,34,35]. All the MDR clones are linked with fluoroquinolone resistance. Most are linked with CTX-M enzymes (i.e., ST131, ST405, ST38, ST648, ST410), and some are linked with carbapenemases (i.e., ST410, ST131, ST405, ST167) [33, 34, 36, 37]. The exact prevalence of ExPEC clones depends on the geographic location, inclusion criteria, sources, and time periods of studies [35].
Extraintestinal Pathogenic Escherichia coli: Epidemic IncF Plasmids
Epidemic IncF plasmids are the most abundant plasmid types among E. coli, especially within ExPEC [38]. IncF plasmids are examples of low copy number, conjugative, narrow host range, epidemic plasmids [4]. These plasmids have been identified in various global ExPEC lineages (among AMR and non-AMR isolates) obtained from humans, animals, and the environment since the 1950s [38]. IncF plasmids contain different replicons (i.e., RepFIA, RepFIIA, RepFIB, RepFIC) [38]. pMLST targets each individual IncF replicon and gives them a number (e.g., F1:A1:B1 etc.) [11, 39].
IncF plasmids are large (> 80 kb), mosaic, and were among the earliest plasmids to be linked with AMR genes [19]. These AMR genes include the following: β-lactamases (e.g., blaCTX-M, blaKPC, blaTEM, blaOXA), plasmid-mediated quinolone resistance determinants (e.g., aac-(6′)-Ib-cr), aminoglycoside-modifying enzymes (including aadA2, aadA5, aph(3′)-Ib, aph(6′)-1d, acc(3′)-IId), and several tet, sul, and dfr genes [40]. The genes are responsible for resistance to penicillins, cephalosporins, carbapenems, aminoglycosides, fluoroquinolones, sulfonamides, tetracyclines, and trimethoprim [40]. IncF plasmids also contain genes for various VAFs that encode bacteriocins, siderophores, cytotoxins, or adhesion factors [8, 38]. ISEcp1 played an important role in the capture and increased expression on blaCTX-Ms [41].
E. coli ST1193 is emerging as an important fluoroquinolone-resistant clone that appeared among E. coli in the late 2000s [42]. Its prevalence has increased from 2012 onwards, especially among fluoroquinolone-resistant isolates. In certain regions, it seems that ST1193 is also emerging as a dominant clone among unselected E. coli collections, mirroring the success of another E. coli clone, namely ST131. Fluoroquinolone-resistant ST1193 is rapidly emerging as a cause of global community-onset urinary tract and bloodstream infections associated with sepsis and hospitalization. ST1193 contains various AMR determinants including several β-lactamases [43]. Carbapenemase genes are currently rare among this global MDR clone. Of special concern is the presence of CTX-M genes which are increasing rapidly among this clone over time [44]. Studies regarding the roles of CTX-M-containing plasmids in ST1193 are currently rare.
The remainder of this article will describe the contributions of IncF epidemic plasmids containing blaCTX-Ms in the success of MDR E. coli ST131 and ST410. We choose these MDR clones because of the following reasons: (i) ST131 is the most common MDR clone among ExPEC globally and linked with FQ-R and CTX-Ms. (ii) ST131 is a truly global clone. (iii) ST410 is the most common carbapenemase ExPEC clone. (iv) IncF plasmids in both clones had been analyzed with long and short read sequencing.
Escherichia coli ST131: Epidemic IncF Plasmids with bla CTX-Ms
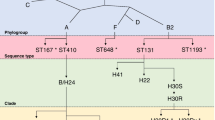
E. coli ST131 is the greatest MDR high-risk global clone of all time [45]. Today, it is a well-established fluoroquinolone-resistant, CTX-M, and carbapenemase-producing global E. coli high-risk clone. ST131 was first described in 2008 [46, 47] and retrospective genomic studies have shown that it was largely responsible for the increase of MDR ExPEC with CTX-Ms during the late 2000s across the globe [48]. ST131 belongs to three clades (A, B, C) and four subclades (C0, C1, C1_M27, C2) [49, 50]. The MDR clade ST131-C with CTX-M β-lactamases dominates the global population structure of this lineage [51].
There are different ST131 clade/CTX-M combinations [52]. CTX-M β-lactamases are extremely rare in clades ST131-A, ST131-B, and ST131-C0. CTX-M-14 is linked with ST131-C1 (approximately 30–40% of C1 is positive for blaCTX-M-14), CTX-M-27 with ST131-C1_M27 (approximately 80–90% of C1_M27 is positive for blaCTX-M-27), and CTX-M-15 with ST131-C2 (approximately 60–70% of C2 is positive for blaCTX-M-15) [51, 53].
Within the ST131-C clades, different IncF plasmids are associated with specific CTX-M enzymes [54]: The F1:A2:B20 plasmids with blaCTX-M-14 are mainly detected in ST131-C1. F1:A2:B20 plasmids with blaCTX-M-27 are found among C1_M27 isolates. The F2:A1:B1 plasmids with blaCTX-M-15 are mainly detected in ST131-C2 [48]. The F1:A2:B20 plasmids often contained the following transposition units: ISEcp1-blaCTX-M-14-IS903B (in ST131-C1) and ISEcp1-blaCTX-M-27-IS903B (in ST131-C1_M27) with the following additional AMR determinants: blaTEM-1, aac(3)-IId, aadA5, aph(3″)-Ib, aph(6)-Id, sul1, sul2, dfrA17, tetA, mph(A) [55]. The F2:A1:B1 plasmids often contained the following transposition unit: ISEcp1-blaCTX-M-15-orf477 with the following additional AMR determinants: blaTEM-1, blaOXA-1, aac(3)-IIa, aadA5, aac(6')-Ib-cr, sul1, dfrA17, and tetA [51, 54, 56]. Both ST131 IncF plasmids (F1:A2:B20 and F2:A1:B1) contained several similar toxin/antitoxin systems, type I restriction‐modification systems, virulence associated genes, and truncated plasmid transfer modules [54, 56].
The ancestral clade, ST131-A, emerged in the mid to late 1800s. In the early to mid 1900s, ST131-A underwent a type 1 pili shift from fimH41 to fimH22/fimH27 to establish ST131-B [45, 57]. During the late 1970s to early 1980s, ST131-B transformed into ST131-C0 by gaining certain QRDR mutations, type 1 pili shift from fimH22/fimH27 to fimH30, and acquiring several integrative genomic elements [57]. During the early to mid 1990s, ST131-C0 gained additional QRDR mutations and split into ST131-C1 and C2 [49, 53, 56].
The role of IncF plasmids in the selective advantage of ST131 are summarized as follows (Fig. 1). F29:B10 plasmids (without blaCTX-Ms) entered the ST131-A lineage around the late 1800s, then moved to ST131-B during the mid 1900s and were lost when ST131-C0 split into C1 and C2. F2:A1:B1 plasmids (without blaCTX-Ms) entered the ST131-B lineage around the 1950s and then moved to ST131-C2 subclade during the early 1900s (Fig. 1). The F2:A1:B1 plasmids lost the B1 replicon and gained several AMR genes (e.g., blaTEM-1, aac(3)-IIa, aadA5, sul1, dfrA17, and tetA) over time. In the mid-late 1990s, F2:A1 plasmids acquired the transposition unit ISEcp1-blaCTX-M-15-orf477, blaOXA-1, and aac(6')-Ib-cr, via IS26-mediated transposition [56].
The F1:A2:B20 plasmids (without blaCTX-Ms) entered the ST131-C1 lineage during the mid 1990s and gained blaTEM-1, aac(3)-IId, aadA5, aph(3″)-Ib, aph(6)-Id, sul1, sul2, dfrA17, and tetA over time (Fig. 1). During the late 1990s, F1:A2:B20 plasmids acquired the transposition unit ISEcp1-blaCTX-M-14-IS903B via IS26-mediated transposition [55]. In the mid-late 2000s, ST131-C1 acquired an additional genomic element (i.e., prophage M27PP1) to become ST131-C1_M27 (Fig. 1) [50]. This was accompanied by a point mutation in blaCTX-M-14 to become blaCTX-M-27. CTX-M-27 has better activity against ceftazidime than CTX-M-14 does [58]. ST131-C1_M27 then increased in frequency among E. coli-producing ESBLs, especially during the mid-late 2010s [30].
Escherichia coli ST410: Epidemic IncF Plasmids with bla CTX-M-15 and bla NDM-5
E. coli ST410 is an emerging carbapenemase-producing global MDR clone [59]. ST410 was initially linked with CTX-Ms in the 2000s. This clone has played an important role in the emergence of CTX-M-15 in the early 2000s [60] and the global distribution of OXA-181 and NDM-5 carbapenemases during the 2010s [61]. ST410 belongs to two clades (ST410-A and ST410-B) and three subclades (ST410-B1, ST410-B2, and ST410-B3) [62].
CTX-M-15 is linked with the ST410-B2 and ST410-B3 subclades [62]. Other types of ESBLs are rare. Within the ST410-B2 subclade, the blaCTX-M-15 is situated on F36:31:A4:B1 plasmids [62]. The CTX-M-15 gene is flanked by IS26 and ISEcp-1 upstream and Tn3, cat, blaOXA-1, aac(6′)-Ib-cr, and IS26 downstream, i.e., IS26-ISEcp-1-blaCTX-M-15-Tn3-cat-blaOXA-1-aac(6′)-Ib-cr-IS26. The following AMR genes were also situated within the F36:31:A4:B1 plasmids, namely aadA2, aac(3′)-II, aadA5, aph(6′)-I, dfrA12, dfrA17, tetA, and sul1. Within the ST410-B3 subclade, the blaCTX-M-15 is situated on F1:A1:B49 plasmids that also contain the following AMR genes: blaNDM-5, blaOXA-1, blaTEM-1, aac(6′)-Ib-cr, aadA2, aadA5, aph(3′)-Ib, aph(6′)-1d, acc(3′)-IId, strA, strB, mph(A), catB4, dfrA12, dfrA17, sul1, sul2, and tetB [62]. Both sets of ST410 IncF plasmids contained several toxin/antitoxin systems, type I restriction‐modification systems, virulence associated genes, and truncated plasmid transfer modules [62].
The ancestral clade (ST410-A) emerged in the mid 1800s. ST410-B1 emerged from ST410-A in the mid 1930s that was associated with a type I pili switch from fimH53 to fimH24. The fluoroquinolone-resistant ST410-B2 subclade evolved from ST410-B1 in the early 1990s. QRDR mutations were acquired via a large homologous recombination event [62].
The role of IncF plasmids in the selective advantage of ST410 is summarized as follows (Fig. 1). The divergence of ST410-B2 from ST410-B1 in the mid-late 1990s correlated with the acquisition of blaCTX-M-15 situated within F36:31:A4:B1 plasmids (Figs. 1, 2). The ST410-B3 subclade evolved from ST410-B2 in 2006, which was accompanied by the replacement of F36:31:A4:B1 plasmids with F1:A1:B49 plasmids harboring blaCTX-M-15 (Figs. 1, 2). The NDM-5 gene was incorporated into the F1:A1:B49 plasmids during the early 2010s, likely via IS26-mediated insertion around 2010 (Figs. 1, 2) [62].
Conclusions
Epidemic IncF plasmids have been pivotal in the selective advantage of certain ExPEC clones such as ST131 and ST410. They offered several advantages to their hosts that allowed the plasmids to coevolve with the bacterial genomes, playing an integral role in the success of their hosts. IncF plasmids are large, mosaic, and often contain various types of AMR genes [8]. These plasmids often contain certain VAFs genes (including bacteriocins, cytotoxins, adhesion factors, and siderophores) [40]. IncF plasmids accommodate various addiction systems (i.e., toxin–antitoxin structures), restriction systems, and truncated transfer systems [63]. As a result of the plasticity of IncF plasmids, they continually undergo extensive rearrangements, especially among the accessory genes (AMR and VAF genes). This has enabled them to coevolve with their hosts by acquiring a wide array of different AMR and VAF genes over time. The presence of AMR genes, VAF genes, addition/restriction systems combined with truncated transfer regions, has led to IncF plasmid persistence and stability with subsequent fixation within certain ExPEC lineages such as E. coli ST131 and ST410.
VAF genes on IncF plasmids do not encode true virulence factors and it seems they are important in the enhanced ecological survival and fitness of hosts [8, 40]. This raises an intriguing issue in that antimicrobial agents will positively select for ExPEC lineages that contain IncF plasmids with AMR genes. If such plasmids also harbor VAFs, positive antimicrobial selection pressures will select for AMR bacteria with enhanced ecological survival and fitness abilities [9]. Such features may also help to explain the plasmid paradox and may be partly responsible for the high prevalence of epidemic IncF plasmids among ExPEC populations obtained from different One Health sources and geographical origins.
One of the best examples of how IncF plasmids have contributed to the success of their hosts occurred during the mid-late 2000s with E. coli ST131 clade C [45, 48]. IncF plasmids have shaped ST131-C and global success. The ST131-C and CTX-M IncF plasmid combination has spread globally and is an important cause of human infections [52]. IncF plasmids entered the ST131 ancestral lineage in the mid 1900s and different clade/CTX-M plasmid combinations coevolved over time [52]. The most successful was the IncF_CTX-M-15/ST131-C2 combination that emerged during the early 2000s and spread rapidly across the globe. This combination is one of the greatest clone/plasmid successes of the millennium [45]. The global spread of IncF_CTX-M-15/ST131-C2 has indirectly led to the increased use of carbapenems with the subsequent increase in carbapenem resistance. It is not a mere coincidence that the global increase of carbapenem-resistant E. coli during the 2000s and 2010s mirrored the global emergence and spread of MDR ST131-C2 with blaCTX-M-15.
The same scenario is currently happening with a different E. coli fluoroquinolone-resistant clone named ST1193 [42]. ST1193 has been rapidly emerging since 2015 as a cause of global community-onset urinary tract and bloodstream infections associated with sepsis and hospitalization. Of special concern is the presence of CTX-M genes which are increasing rapidly within this clone over time [44].
E. coli ST410 is another example of how specific IncF plasmids promoted the success of certain subclades within this lineage. Specific ST410-B3 IncF plasmids that contained blaCTX-M-15 gradually incorporated the NDM-5 carbapenemase genes into existing IncF platforms providing an additional positive selective advantage that now also includes the carbapenems (Fig. 2). The “stepwise” acquisition of initially CTX-M genes, followed by gaining carbapenemase genes by ST410, is shared with certain K. pneumoniae MDR high-risk clones (e.g., ST147, ST307) [64].
The use of antimicrobial agents will continue to create positive selection pressures that enhance the risks for the continuous emergence of MDR high-risk clone/IncF plasmid combinations. ST131-C2/IncF_CTX-M-15 and ST410-B3/IncF_CTX-M-15_NDM-5 are recent examples. A “plasmid-replacement” clade scenario occurred in the histories of ST131 and ST410 as different subclades gained different AMR genes on different IncF platforms. The reasons for the IncF replacements and associations between certain ST131/ST410 subclades and specific IncF plasmid types are unknown. Research projects aimed at investigating the important features responsible for the success of such combinations need to be funded and urgently addressed. Such information will aid in designing management and prevention strategies for clade/plasmid combinations. These projects will also serve as models to predict the future emergence of successful clones/plasmid combinations among clinically relevant Gram-negative bacteria.
As the medical community faces the potential of a post-antibiotic era, fully understanding the underlying reasons for the emergence and spread of successful MDR clade/plasmid combinations is critical to developing means for slowing down the dissemination of plasmid-encoded AMR genes. The medical community can ill afford to ignore the continuous emergence and spread of globally successful MDR high-risk E. coli clones/plasmid combinations.
References
Baquero F. Threats of antibiotic resistance: an obliged reappraisal. Int Microbiol. 2021;24(4):499–506.
Antimicrobial RC. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–55.
Baker S, Thomson N, Weill FX, Holt KE. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science. 2018;360(6390):733–8.
Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28(3):565–91.
Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31(4). https://doi.org/10.1128/CMR.00088-17.
Baquero F, Martinez JL, Lanza VF, et al. Evolutionary pathways and trajectories in antibiotic resistance. Clin Microbiol Rev. 2021;34(4):e0005019.
Rodriguez-Beltran J, DelaFuente J, Leon-Sampedro R, MacLean RC, San MA. Beyond horizontal gene transfer: the role of plasmids in bacterial evolution. Nat Rev Microbiol. 2021;19(6):347–59.
Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303(6–7):298–304.
Baquero F, Martinez JL, Lanza VF, et al. Evolutionary pathways and trajectories in antibiotic resistance. Clin Microbiol Rev. 2021:e0005019.
Carattoli A. Plasmids in Gram negatives: molecular typing of resistance plasmids. Int J Med Microbiol. 2011;301(8):654–8.
Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124.
Garcillan-Barcia MP, Alvarado A, de la Cruz F. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol Rev. 2011;35(5):936–56.
Guglielmini J, de la Cruz F, Rocha EP. Evolution of conjugation and type IV secretion systems. Mol Biol Evol. 2013;30(2):315–31.
Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010;74(3):434–52.
Bottery MJ. Ecological dynamics of plasmid transfer and persistence in microbial communities. Curr Opin Microbiol. 2022;68:102152.
Jain A, Srivastava P. Broad host range plasmids. FEMS Microbiol Lett. 2013;348(2):87–96.
Harrison E, Brockhurst MA. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012;20(6):262–7.
Brockhurst MA, Harrison E. Ecological and evolutionary solutions to the plasmid paradox. Trends Microbiol. 2022;30(6):534–43.
Helinski DR. A brief history of plasmids. EcoSal Plus. 2022;10(1):eESP00282021.
Pitout JD. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol. 2012;3:9.
Laupland KB, Church DL. Population-based epidemiology and microbiology of community-onset bloodstream infections. Clin Microbiol Rev. 2014;27(4):647–64.
Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014;20(9):821–30.
Bonten M, Johnson JR, van den Biggelaar AHJ, et al. Epidemiology of Escherichia coli bacteremia: a systematic literature review. Clin Infect Dis. 2021;72(7):1211–9.
Naylor NR, Pouwels KB, Hope R, et al. The health and cost burden of antibiotic resistant and susceptible Escherichia coli bacteraemia in the English hospital setting: a national retrospective cohort study. PLoS ONE. 2019;14(9):e0221944.
Leger A, Lambraki I, Graells T, et al. Characterizing social-ecological context and success factors of antimicrobial resistance interventions across the One Health spectrum: analysis of 42 interventions targeting E. coli. BMC Infect Dis. 2021;21(1):873.
Pitout JD. Extraintestinal pathogenic Escherichia coli: an update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev Anti Infect Ther. 2012;10(10):1165–76.
Peirano G, Pitout JD. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents. 2010;35(4):316–21.
Pitout JD, Chan WW, Church DL. Tackling antimicrobial resistance in lower urinary tract infections: treatment options. Expert Rev Anti Infect Ther. 2016;14(7):621–32.
Fuzi M, Rodriguez Bano J, Toth A. Global evolution of pathogenic bacteria with extensive use of fluoroquinolone agents. Front Microbiol. 2020;11:271.
Peirano G, Pitout JDD. Extended-spectrum beta-lactamase-producing enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs. 2019;79(14):1529–41.
Pitout JDD. Population dynamics of Escherichia coli causing bloodstream infections over extended time periods. mSphere. 2021;6(6):e0095621.
Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–84.
Peirano G, Chen L, Nobrega D, et al. Genomic epidemiology of global carbapenemase-producing Escherichia coli, 2015–2017. Emerg Infect Dis. 2022;28(5):924–31.
Cummins EA, Snaith AE, McNally A, Hall RJ. The role of potentiating mutations in the evolution of pandemic Escherichia coli clones. Eur J Clin Microbiol Infect Dis. 2021. https://doi.org/10.1007/s10096-021-04359-3.
Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD. Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin Microbiol Rev. 2019;32(3):e00135-18.
Peirano G, Bradford PA, Kazmierczak KM, et al. Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg Infect Dis. 2014;20(11):1928–31.
Peirano G, van der Bij AK, Freeman JL, et al. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum beta-lactamases: global distribution of the H30-Rx sublineage. Antimicrob Agents Chemother. 2014;58(7):3762–7.
Johnson TJ, Nolan LK. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev. 2009;73(4):750–74.
Carattoli A, Zankari E, Garcia-Fernandez A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–903.
Johnson TJ. Role of plasmids in the ecology and evolution of "high-risk" extraintestinal pathogenic Escherichia coli clones. EcoSal Plus. 2021;9(2). https://doi.org/10.1128/ecosalplus.ESP-0013-2020.
Poirel L, Decousser JW, Nordmann P. Insertion sequence ISEcp1B is involved in expression and mobilization of a bla(CTX-M) beta-lactamase gene. Antimicrob Agents Chemother. 2003;47(9):2938–45.
Pitout JDD, Peirano G, Chen L, DeVinney R, Matsumura Y. Escherichia coli ST1193: following in the footsteps of E. coli ST131. Antimicrob Agents Chemother. 2022;66(7):e00511-22.
Johnson TJ, Elnekave E, Miller EA, et al. Phylogenomic analysis of extraintestinal pathogenic Escherichia coli sequence type 1193, an emerging multidrug-resistant clonal group. Antimicrob Agents Chemother. 2019;63(1):e01913-18.
Peirano G, Matsumara Y, Nobrega D, DeVinney R, Pitout J. Population-based epidemiology of Escherichia coli ST1193 causing blood stream infections in a centralized Canadian region. Eur J Clin Microbiol Infect Dis. 2021. https://doi.org/10.1007/s10096-021-04373-5.
Pitout JDD, Finn TJ. The evolutionary puzzle of Escherichia coli ST131. Infect Genet Evol. 2020;81:104265.
Coque TM, Novais A, Carattoli A, et al. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis. 2008;14(2):195–200.
Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother. 2008;61(2):273–81.
Pitout JD, DeVinney R. Escherichia coli ST131: a multidrug-resistant clone primed for global domination. F1000Res. 2017;6(F1000 Faculty Rev):195. https://doi.org/10.12688/f1000research.10609.1.
Petty NK, Ben Zakour NL, Stanton-Cook M, et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci USA. 2014;111(15):5694–9.
Matsumura Y, Pitout JD, Gomi R, et al. Global Escherichia coli sequence type 131 clade with blaCTX-M-27 gene. Emerg Infect Dis. 2016;22(11):1900–7.
Decano AG, Downing T. An Escherichia coli ST131 pangenome atlas reveals population structure and evolution across 4,071 isolates. Sci Rep. 2019;9(1):17394.
Mathers AJ, Peirano G, Pitout JD. Escherichia coli ST131: the quintessential example of an international multiresistant high-risk clone. Adv Appl Microbiol. 2015;90:109–54.
Peirano G, Lynch T, Matsumara Y, et al. Trends in population dynamics of Escherichia coli sequence type 131, Calgary, Alberta, Canada, 2006–2016(1). Emerg Infect Dis. 2020;26(12):2907–15.
Johnson TJ, Danzeisen JL, Youmans B, et al. Separate F-type plasmids have shaped the evolution of the H30 subclone of Escherichia coli sequence type 131. mSphere. 2016;1(4):e00121-16.
Hayashi M, Matsui M, Sekizuka T, et al. Dissemination of IncF group F1:A2:B20 plasmid-harbouring multidrug-resistant Escherichia coli ST131 before the acquisition of bla(CTX-M) in Japan. J Glob Antimicrob Resist. 2020;23:456–65.
Stoesser N, Sheppard AE, Pankhurst L, et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio. 2016;7(2):e02162.
Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, et al. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio. 2016;7(2):e00347–16.
Bonnet R, Recule C, Baraduc R, et al. Effect of D240G substitution in a novel ESBL CTX-M-27. J Antimicrob Chemother. 2003;52(1):29–35.
Roer L, Overballe-Petersen S, Hansen F, et al. Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere. 2018;3(4):e00337-18.
Peirano G, van der Bij AK, Gregson DB, Pitout JD. Molecular epidemiology over an 11-year period (2000 to 2010) of extended-spectrum beta-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J Clin Microbiol. 2012;50(2):294–9.
Feng Y, Liu L, Lin J, et al. Key evolutionary events in the emergence of a globally disseminated, carbapenem resistant clone in the Escherichia coli ST410 lineage. Commun Biol. 2019;2:322.
Chen L, Peirano G, Kreiswirth BN, Devinney R, Pitout JDD. Acquisition of genomic elements were pivotal for the success of Escherichia coli ST410. J Antimicrob Chemother. 2022;77(12):3399–3407.
Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):736–55.
Peirano G, Chen L, Kreiswirth BN, Pitout JDD. Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob Agents Chemother. 2020;64(10):e01148-20.
Acknowledgements
Funding
This work was supported by research grants from the JPIAMR/Canadian Institute Health Research program (#10016015) and National Institute of Health (#10028552). The study is in part supported by NIAID grant R01AI090155. No funding or sponsorship was received for the publication of this article.
Author Contributions
Johann Pitout and Liang Chen conceived the idea, designed the outlay of this review article, and performed the literature search. Johann Pitout wrote the first draft. Liang Chen added sections and approved the final submitted version.
Disclosures
Johann Pitout and Liang Chen confirm that they have no conflicts of interest to declare.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Data Availability
Data sharing is not applicable to this article as no data sets were generated or analyzed during the current study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Pitout, J.D.D., Chen, L. The Significance of Epidemic Plasmids in the Success of Multidrug-Resistant Drug Pandemic Extraintestinal Pathogenic Escherichia coli. Infect Dis Ther 12, 1029–1041 (2023). https://doi.org/10.1007/s40121-023-00791-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40121-023-00791-4