Abstract
Introduction
To promote judicious prescribing of methicillin-resistant Staphylococcus aureus (MRSA)-active therapy for skin and soft tissue infections (SSTI), we previously developed an MRSA risk assessment tool. The objective of this study was to validate this risk assessment tool internationally.
Methods
A multicenter, prospective cohort study of adults with purulent SSTI was performed at seven international sites from July 2016 to March 2018. Patient MRSA risk scores were computed as follows: MRSA infection/colonization history (2 points); previous hospitalization, previous antibiotics, chronic kidney disease, intravenous drug use, human immunodeficiency virus/acquired immunodeficiency syndrome (HIV/AIDS), diabetes with obesity (1 point each). Predictive performance of MRSA surveillance percentage, MRSA risk score, and estimated MRSA probability (surveillance percentage adjusted by risk score) were quantified using the area under the receiver operating characteristic curves (aROC) and compared. Performance characteristics of different risk score thresholds across varying baseline MRSA prevalence were examined.
Results
Two hundred three patients were included. Common SSTI were wounds (28.6%), abscess (25.1%), and cellulitis with abscess (20.7%). Patients with higher risk scores were more likely to have MRSA (P < 0.001). The MRSA risk score aROC (95%CI) [0.748 (0.678–0.819)] was significantly greater than MRSA surveillance percentage [0.646 (0.569–0.722)] (P = 0.016). Estimated MRSA probability aROC [0.781 (0.716–0.845)] was significantly greater than surveillance percentage (P < 0.001) but not the risk score (P = 0.192). The estimated negative predictive value (NPV) of an MRSA score ≥ 1 (i.e., a score of 0) was greater than 90% when MRSA prevalence was 30% or less.
Conclusion
The MRSA risk score and estimated MRSA probability were significantly more predictive of MRSA compared with surveillance percentage. An MRSA risk score of zero had high predictive value and could help avoid unnecessary empiric MRSA coverage in low-acuity patients. Further study, including impact of such risk assessment tools on prescribing patterns and outcomes are required before implementation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Stratifying patients with skin and soft tissue infections (SSTI) based on their risk for methicillin-resistant Staphylococcus aureus (MRSA) has the potential to improve anti-MRSA antibiotic prescribing and promote good patient and public health outcomes. |
This study evaluated the predictive performance of various MRSA risk assessment approaches based on previously developed MRSA SSTI risk assessment tools in an international cohort of patients. |
The MRSA risk score, with or without inclusion of nationally reported MRSA surveillance percentage, was significantly more predictive of MRSA than the national surveillance percentage alone. |
An MRSA risk score of zero had high predictive value for a non-MRSA infection, demonstrating that this tool could help avoid unnecessary empiric MRSA coverage in low-acuity patients. |
Introduction
Skin and soft tissue infections (SSTI) are among the most common infections encountered in community and healthcare settings. [1] Worldwide, bacterial skin diseases, which includes cellulitis and abscesses, accounted for nearly 267 million new cases and contributed to an increase in years lived with disability. [2] A primary SSTI antibiotic treatment consideration is whether methicillin-resistant Staphylococcus aureus (MRSA) coverage is necessary. The rate of MRSA SSTI increased at an alarming rate in the early 2000s in the USA, likely contributing to increased prescription of MRSA-active antibiotics for skin infections [3,4,5]. Increasing importance of MRSA among SSTI has also been reported worldwide. However, MRSA prevalence differs between regions and countries [6,7,8,9,10]. Prevalence of MRSA ranges from less than 1% in some European countries to upwards of 60% in certain parts of South America, Asia, and the USA [11,12,13]. With approximately 50% MRSA prevalence in many regions, local surveillance data and antibiograms become less useful for clinical decision-making. Consequently, SSTI prescribing practices vary widely, whereby both under- and over-prescribing of MRSA-active antibiotics occurs frequently [5, 14,15,16,17]. This antibiotic misuse has important implications for both patient outcomes and public health.
The ubiquitous nature of skin infections coupled with the challenges associated with empiric antibiotic selection underscore the critical need for improved clinical decision support tools to facilitate antimicrobial stewardship [18]. One potential approach to address this need is individual patient risk assessment to identify those at low or high risk of having MRSA SSTI. Numerous MRSA skin infection risk factors have been reported in the literature [19,20,21,22,23,24,25,26,27,28,29,30]. To aid in clinical application of these risk factors, we previously developed a risk scoring tool that combined data from previous literature with individual patient data from a single health system in southeastern Michigan [31]. Given the widespread variability in MRSA prevalence and regional differences in MRSA epidemiology, the objectives of this study were to validate this risk scoring tool in an international cohort.
Methods
Study Design and Population
This was a multicenter, observational, prospective cohort study of adults with SSTI presenting to seven international hospital or clinic sites from July 2016 to March 2018. Sites included seven university-affiliated clinics and/or hospitals from China, Italy, Mexico, Russia, Singapore, and the UK. Sites were selected to be as geographically diverse as possible and included investigators who regularly treat patients with SSTI and volunteered to participate in the study as an investigator. Patients who did not have a culture obtained, those with osteomyelitis and/or septic arthritis, an SSTI secondary to bite wounds, odontogenic infection, and those who were pregnant or prisoners were excluded. There were no missing data given the prospective nature of the data collection. This study was approved by the Wayne State University institutional review board and waiver of informed consent was granted. The study was also approved by a human subject research committee at each study site when necessary, waiver of informed consent granted, and was conducted in accordance with the principles of the Declaration of Helsinki of 1964, and it's later amendments.
Patient Data Elements and Collection
Patients with SSTI were identified for inclusion by clinician investigators at each study site. Patient demographic and clinical data were obtained from patient interviews and health records and electronically entered into a structured data collection form within Research Electronic Data Capture (REDCap, Vanderbilt University) [32]. SSTI definition and type was determined by the treating clinician and investigator at the time of study inclusion. Data included social history, past medical history, and comorbid conditions previously identified as MRSA risk factors, including those used to compute our previously derived MRSA SSTI risk score [31]. Tissue or wound cultures obtained as part of routine care and results that were available at study enrollment were collected.
Individual Patient MRSA Risk Assessment
The MRSA risk for each patient was quantified using our previously published MRSA risk score [31]. Individual MRSA risk scores were computed as follows: MRSA infection/colonization history (2 points), hospitalization in the previous year (1 point), antibiotic exposure in the previous 6 months (1 point), chronic kidney disease (1 point), intravenous drug use (1 point), human immunodeficiency virus (HIV) infection or acquired immunodeficiency syndrome (AIDS) (1 point), diabetes with obesity (1 point). Fagan’s nomogram and the likelihood ratio of the MRSA scores from the derivation cohort were then used to convert national (country-level) surveillance MRSA percentage (prior probability) into an estimated MRSA probability for each included patient (posterior probability) [33]. Country-level surveillance MRSA percentages were obtained from the European Centre for Disease Prevention and Control and World Health Organization [11, 12].
Data Analysis
The primary analysis evaluated the predictive performance of three MRSA risk assessment approaches: (1) national surveillance MRSA percentage alone, (2) MRSA risk score alone, and (3) MRSA probability estimate (combination of national surveillance and risk score). Predictive performance of each approach was quantified by area under the receiver operating characteristics curve (aROC) along with a 95% confidence interval. The aROC of each risk assessment approach was compared using the Hanley and McNeil method [34].
The secondary analyses evaluated the ability of candidate MRSA risk score thresholds to identify patients at lowest and highest MRSA risk. Observed performance characteristics, including aROC, sensitivity, specificity, negative predictive value (NPV), and positive predictive value (PPV) were computed with 95% confidence intervals for each MRSA risk score threshold with at least 15 patients (e.g., 15 patients with a risk score ≥ 4). Estimated NPV and PPV for each threshold was also computed across hypothetical MRSA prevalence scenarios that were different from what was observed in the international cohort using conditional probability equations [35].
All statistical tests were two-sided; P values ≤ 0.05 were considered statistically significant. Analyses were performed using SPSS Statistics, IBM SPSS software, version 26.0 (IBM Corp., Armonk, NY). No formal sample size calculation was used and a convenience sample of patients was sought.
Results
Two hundred three patients across seven international sites were included. A full description of the cohort is displayed in Supplementary Table 1. The majority of patients (59.6%) were male and either white (82.3%) or Asian (12.3%). The median (IQR) age was 54 (35–67) years. The most common infection types were wound (28.6%), abscess (25.1%), and cellulitis with abscess (20.7%). The most common MRSA risk factors observed were systemic antibiotic exposure in the previous 6 months (45.8%), hospitalization in the previous year (42.4%), diabetes mellitus (30.5%), obesity (23.2%), diabetes with obesity (11.8%), previous abscess (15.8%), MRSA colonization or infection history (12.3%), and intravenous drug use (11.3%). Overall, one-third (33%) of patients had culture-positive MRSA SSTI. Nationally reported surveillance MRSA percentages and observed MRSA percentages at each study site are shown in Fig. 1. Observed MRSA percentages ranged from 10% in Beijing, China to 58.8% in Mexico City, Mexico. The number of patients from each site also ranged from 10 patients in Beijing, China to 50 patients from both St. Petersburg, Russia and Winchester, UK, respectively. In general, study sites with lower national surveillance MRSA percentages saw correspondingly lower observed MRSA percentages. The MRSA risk score distribution and observed MRSA percentages by risk score values are shown in Fig. 2. The majority (58.2%) of patients had a risk score of 0 or 1. The median (IQR) MRSA risk score of the entire cohort was 1 (0–2). As the MRSA risk score increased, the proportion of patients with MRSA increased significantly (P < 0.001).
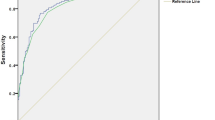
The predictive performances of the national surveillance MRSA percentage, the MRSA risk score, and the MRSA probability estimate (surveillance MRSA percentage adjusted by the MRSA risk score) are presented in Table 1. The predictive performance of surveillance MRSA percentage was significantly better than random chance, with an AUC (95% CI) of 0.646 (0.569–0.722). The MRSA risk score performed significantly better than surveillance percentage, with an AUC (95% CI) of 0.748 (0.678–0.819) (P = 0.016). The MRSA probability estimate was also significantly more predictive relative to surveillance percentage, with an AUC (95% CI) of 0.781 (0.716–0.845) (P < 0.001). However, no significant difference in aROC was observed between the MRSA risk score and MRSA probability estimate.
The performance characteristics of various risk score thresholds for predicting MRSA are presented in Table 2. Overall predictive performance, as measured by aROC, was maximized at a risk score threshold ≥ 2 (AUC 0.733, 95% CI 0.658–0.808). Sensitivity and negative predictive value were maximized at a risk score threshold of ≥ 1 (sensitivity 91.0%, 95% CI 84.2–97.9; NPV 89.8%, 95% CI 82.1–97.5). Ten percent of subjects with a risk score of 0 had MRSA (Fig. 2). Sensitivity and negative predictive value progressively decreased as risk score thresholds increased. Specificity and positive predictive value were maximized at a risk score threshold of ≥ 4 (specificity 94.9%, 95% CI 91.1–98.6; PPV 63.2%, 95% CI 41.5–84.8). Positive predictive value of risk score thresholds of ≥ 2, ≥ 3, and ≥ 4 were similar and ranged from 54.1% to 63.2%. Among patients with a risk score ≥ 2, 57.6% had MRSA (Fig. 2).
The estimated positive and negative predictive values of an MRSA risk score ≥ 1 across various hypothetical MRSA prevalence settings are displayed in Table 3. The estimated NPV of an MRSA score ≥ 1 (i.e., a risk score of 0) was in excess of 90% when MRSA prevalence was 30% or less. When MRSA prevalence was > 50%, the NPV of a risk score ≥ 1 was below 80%. The estimated PPV of an MRSA score ≥ 1 was poor until MRSA prevalence was very high (> ~70%). The estimated positive and negative predictive values of MRSA risk score thresholds are displayed in Supplementary Table 2. These thresholds generally had better estimated PPV than a threshold of ≥ 1 but all candidate thresholds had a PPV ≤ 76% at an MRSA prevalence of 50%.
Discussion
This study sought to internationally validate an MRSA risk scoring tool developed from previously published literature and individual patient data from a single US health system [31]. The MRSA risk score demonstrated fair predictive performance in an international cohort derived from seven study sites. The aROC score (95% CI) of 0.748 (0.678–0.819) was greater than the original derivation cohort 0.601 (95% CI 0.521–0.681). The score was significantly more predictive of MRSA relative to national surveillance MRSA percentages alone. Using Fagan’s nomogram, the score was also able to generate individual patient-predicted MRSA probabilities from the MRSA surveillance percentages. These MRSA probability estimates also demonstrated fair predictive performance and was a significant improvement from MRSA surveillance percentages alone. Collectively, these approaches demonstrate that the use of individual patient characteristics can better predict MRSA SSTI than the use of general surveillance data alone. This demonstrates the potential for similar approaches to be used in clinical practice when a suitable risk stratification model is available. This could include electronic health record-based clinical decision support tools incorporating Fagan’s nomogram or simple bedside scoring systems such as the MRSA score.
No significant difference in predictive performances between the MRSA probability estimates and the MRSA risk scores was observed. This is a noteworthy finding in the context of the widely varying MRSA prevalence observed in our study and in clinical practice. Unlike the probability estimate, the MRSA risk score only accounts for individual patient clinical characteristics and does not account for national MRSA prevalence. Knowledge of local MRSA prevalence would theoretically improve the ability to predict MRSA presence. Although there was no significant difference, the aROC for estimated MRSA probability was numerically greater than the aROC for the MRSA risk score. It is unclear whether this convenience sample was large enough to exclude a meaningful difference in aROC but, at minimum, it appears that accounting for national MRSA prevalence did not reduce the ability of the risk score to predict MRSA. This finding indicates that the MRSA risk score may be reasonably applied without knowledge of MRSA prevalence in settings where it is either unknown, or computation of estimated MRSA probability estimate is difficult or impossible.
Despite less than optimal predictive performance, these risk scoring approaches were able to identify patients at lowest and highest risk of MRSA infection comparably to other risk scores [36]. Methicillin-resistant S. aureus was observed in 10% of patients with a risk score of zero and nearly 60% in those with a risk score of two or more. An MRSA risk score of zero had an NPV in excess of 90% even when hypothetical MRSA prevalence was 30%, and an NPV in excess of 80% up to a hypothetical MRSA prevalence of 50%. This hypothetical MRSA prevalence range encompasses all but two of the study sites from this international cohort, suggesting this MRSA prevalence range is common. Risk of MRSA, along with patient clinical status and disease severity, may be used to better inform diagnostic and antibiotic treatment decisions. Clinicians may be able to avoid MRSA coverage in low-acuity patients with an MRSA risk score of zero. The majority of patients in this and similar studies are at low MRSA risk and this could greatly reduce the number of patients receiving unnecessary MRSA coverage. [16, 36] Such an approach would prevent unnecessary MRSA coverage in approximately 25% of the patients in this study. It would only result in failure to cover MRSA when necessary in 3% of the patients in this study. Although failing to provide in vitro active therapy is never desirable, studies suggest that a 3% rate of under-prescribing is less frequent than estimates from a cohort of patients with abscess receiving antibiotics [5].
These findings should be interpreted in the context of several considerations and limitations. Although this study comprehensively collected MRSA risk factors through health records and patient interviews, it is possible that some clinical data or risk factors were missed or were present too infrequently to assess (e.g., recent travel from high-prevalence area). Prospective MRSA colonization screenings were not conducted at all study sites and thus, it is possible some colonized patients were considered non-colonized [37]. However, MRSA infection/colonization history was strongly associated with MRSA SSTI in this study, thus suggesting this potential non-differential misclassification bias did not substantially alter the findings. It is also important to note that although inclusion of multiple study sites across six countries promotes external validity of the findings, the generalizability is not without several limitations. Firstly, this study focused primarily on purulent SSTI to facilitate organism identification. Therefore, these findings may not apply to patients with non-purulent skin infections. Secondly, the majority of patients were also managed in various hospital settings rather than clinics, thereby potentially limiting applicability in the ambulatory and community settings. It is unclear what proportion of these infections were healthcare-associated versus community acquired. Patient management differences across sites could also introduce selection bias into the findings. Finally, the limited patient numbers from some study sites may have limited the ability to fully control for site effects when deriving an adjusted risk score. Despite this, the study sites that were controlled for did not result in exclusion of risk factors from the initial risk score, suggesting that site effect would not substantially change the most important risk factors. Lastly, although country-level surveillance MRSA percentages were obtained from the European Centre for Disease Prevention and Control and World Health Organization, this may not be the best method to estimate true local prevalence and it is unclear if more granular local data would improve or impede the ability of these methods to predict MRSA.
Conclusions
In conclusion, an MRSA SSTI risk assessment with an individual patient approach using previously published MRSA risk factors with or without knowledge of national MRSA surveillance percentage was significantly more predictive of MRSA SSTI than MRSA surveillance percentages alone. Such risk assessment approaches may be simply administered at the point of care (risk score) or through an electronic clinical decision support application (risk score or MRSA probability estimate). It is unclear at this time how such risk scoring approaches would affect antibiotic prescribing patterns or patient outcomes in SSTI. Future research to evaluate this or other MRSA risk scoring approaches in SSTI antimicrobial stewardship should be conducted prior to widespread clinical implementation.
References
Kaye KS, Petty LA, Shorr AF, Zilberberg MD. Current epidemiology, etiology, and burden of acute skin infections in the United States. Clin Infect Dis. 2019;68(Suppl 3):S193–9.
Disease GBD, Injury I, Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1789–858.
Stryjewski ME, Chambers HF. Skin and soft-tissue infections caused by community-acquired methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008;46(Suppl 5):S368–77.
Johnson JK, Khoie T, Shurland S, Kreisel K, Stine OC, Roghmann MC. Skin and soft tissue infections caused by methicillin-resistant Staphylococcus aureus USA300 clone. Emerg Infect Dis. 2007;13(8):1195–200.
Pallin DJ, Camargo CA Jr, Schuur JD. Skin infections and antibiotic stewardship: analysis of emergency department prescribing practices, 2007–2010. West J Emerg Med. 2014;15(3):282–9.
Klein S, Menz MD, Zanger P, Heeg K, Nurjadi D. Increase in prevalence of PVL and clonal shift in community onset-MRSA causing skin and soft tissue infections in the Rhine-Neckar-region, Germany, 2012–2016. Int J Antimicrob Agents. 2018;53:261–7.
Macmorran E, Harch S, Athan E, et al. The rise of methicillin resistant Staphylococcus aureus: now the dominant cause of skin and soft tissue infection in Central Australia. Epidemiol Infect. 2017;145(13):2817–26.
Li X, Chen Y, Gao W, Ouyang W, Wei J, Wen Z. Epidemiology and outcomes of complicated skin and soft tissue infections among inpatients in southern China from 2008 to 2013. PLoS One. 2016;11(2): e0149960.
Dozois A, Thomsen I, Jimenez-Truque N, et al. Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus among skin and soft tissue infections in an emergency department in Guyana. Emerg Med J EMJ. 2015;32(10):800–3.
Diekema DJ, Pfaller MA, Shortridge D, Zervos M, Jones RN. Twenty-year trends in antimicrobial susceptibilities among Staphylococcus aureus from the SENTRY antimicrobial surveillance program. Open Forum Infect Dis. 2019;6(Suppl 1):S47-s53.
(ECDC) ECfDPaC. Antimicrobial resistance surveillance in Europe 2014. https://ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2014. Accessed Feb 13.
(WHO) WHO. Antimicrobial resistance: global report on surveillance 2014. http://www.who.int/drugresistance/documents/surveillancereport/en/. Accessed Feb 13.
(CDC) CfDCaP. National healthcare safety network healthcare-associated infection antibiotic resistance data. https://gis.cdc.gov/grasp/PSA/MapView.html. Accessed Feb 13.
Mistry RD, Shapiro DJ, Goyal MK, et al. Clinical management of skin and soft tissue infections in the U.S. emergency departments. West J Emerg Med. 2014;15(4):491–8.
Meddles-Torres C, Hu S, Jurgens C. Changes in prescriptive practices in skin and soft tissue infections associated with the increased occurrence of community acquired methicillin resistant Staphylococcus aureus. J Infect Public Health. 2013;6(6):423–30.
Sutton JD, Carico R, Burk M, et al. Inpatient management of uncomplicated skin and soft tissue infections in 34 veterans affairs medical centers: a medication use evaluation. Open Forum Infect Dis. 2020;7(1):ofz554.
Rhoads JLW, Willson TM, Sutton JD, Spivak ES, Samore MH, Stevens VW. Epidemiology, disposition and treatment of ambulatory Veterans with skin and soft tissue infections. Clin Infect Dis. 2020;72:675–81.
Gibbons JA, Smith HL, Kumar SC, et al. Antimicrobial stewardship in the treatment of skin and soft tissue infections. Am J Infect Control. 2017;45(11):1203–7.
Moran GJ, Krishnadasan A, Gorwitz RJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355(7):666–74.
Frazee BW, Salz TO, Lambert L, Perdreau-Remington F. Fatal community-associated methicillin-resistant Staphylococcus aureus pneumonia in an immunocompetent young adult. Ann Emerg Med. 2005;46(5):401–4.
Miller LG, Perdreau-Remington F, Bayer AS, et al. Clinical and epidemiologic characteristics cannot distinguish community-associated methicillin-resistant Staphylococcus aureus infection from methicillin-susceptible S. aureus infection: a prospective investigation. Clin Infect Dis. 2007;44(4):471–82.
Stenstrom R, Grafstein E, Romney M, et al. Prevalence of and risk factors for methicillin-resistant Staphylococcus aureus skin and soft tissue infection in a Canadian emergency department. CJEM. 2009;11(5):430–8.
Szumowski JD, Wener KM, Gold HS, et al. Methicillin-resistant Staphylococcus aureus colonization, behavioral risk factors, and skin and soft-tissue infection at an ambulatory clinic serving a large population of HIV-infected men who have sex with men. Clin Infect Dis. 2009;49(1):118–21.
Gadepalli R, Dhawan B, Kapil A, et al. Clinical and molecular characteristics of nosocomial meticillin-resistant Staphylococcus aureus skin and soft tissue isolates from three Indian hospitals. J Hosp Infect. 2009;73(3):253–63.
Maree CL, Eells SJ, Tan J, et al. Risk factors for infection and colonization with community-associated methicillin-resistant Staphylococcus aureus in the Los Angeles County jail: a case-control study. Clin Infect Dis. 2010;51(11):1248–57.
Khawcharoenporn T, Tice AD, Grandinetti A, Chow D. Risk factors for community-associated methicillin-resistant Staphylococcus aureus cellulitis—and the value of recognition. Hawaii Med J. 2010;69(10):232–6.
Weiss C, Kaminsky P, Boggs J, Ley C. Skin and soft-tissue infections in suburban primary care: epidemiology of methicillin-resistant Staphylococcus aureus and observations on abscess management. BMC Res Notes. 2011;4:33.
Achiam CC, Fernandes CM, McLeod SL, et al. Methicillin-resistant Staphylococcus aureus in skin and soft tissue infections presenting to the Emergency Department of a Canadian Academic Health Care Center. Eur J Emerg Med Off J Eur Soc Emerg Med. 2011;18(1):2–8.
Vayalumkal JV, Suh KN, Toye B, Ramotar K, Saginur R, Roth VR. Skin and soft tissue infections caused by methicillin-resistant Staphylococcus aureus (MRSA): an affliction of the underclass. CJEM. 2012;14(6):335–43.
Chou YH, Lee MS, Lin RY, Wu CY. Risk factors for methicillin-resistant Staphylococcus aureus skin and soft-tissue infections in outpatients in Taiwan. Epidemiol Infect. 2015;143(4):749–53.
Claeys KC, Zasowski EJ, Lagnf AM, Levine DP, Davis SL, Rybak MJ. Novel application of published risk factors for methicillin-resistant S. aureus in acute bacterial skin and skin structure infections. Int J Antimicrob Agents. 2018;51(1):43–6.
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–81.
Fagan TJ. Letter: nomogram for Bayes theorem. N Engl J Med. 1975;293(5):257.
Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148(3):839–43.
Molinaro AM. Diagnostic tests: how to estimate the positive predictive value. Neurooncol Pract. 2015;2(4):162–6.
Zilberberg MD, Chaudhari P, Nathanson BH, et al. Development and validation of a bedside risk score for MRSA among patients hospitalized with complicated skin and skin structure infections. BMC Infect Dis. 2012;12:154.
Acquisto NM, Bodkin RP, Brown JE, et al. MRSA nares swab is a more accurate predictor of MRSA wound infection compared with clinical risk factors in emergency department patients with skin and soft tissue infections. Emerg Med J EMJ. 2018;35(6):357–60.
Acknowledgements
We thank the participants of this study.
Funding
This study was funded by an investigator-initiated grant from Bayer AG.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contributions
All authors conceived the study, contributed to the study design, were involved in interpretation of the data, revised the manuscript, and approved the final version before submitting. EJZ, KCC, MJR obtained funding. MD, SS, MB, AC, NK, AK, APC, LM, and BC collected the data. EJZ, TDT, and MJR performed statistical analysis. EJZ wrote the first draft of the report, and designed the tables and figures. The corresponding author confirms that he had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Disclosures
KC has received grant funding from Merck & Co, Kenilworth, NJ, the Centers for Disease Control and Prevention (CDC), and Veterans Affairs Health Services Research Merit and honoraria from BioFire Diagnostics, Salt Lake City, Utah. MB has received grant funding from SOBI, MSD, Pfizer, and Shionogi, consulting fees from SOBI, Roche, Menarini, honoraria from MSD, Pfizer, Shionogi, SOBI, and Menarini. NK has received honoraria from Merck and MSD. AK has received honoraria from Amgen and Pfizer, support to attend meetings from MSD and Sumitomo Dainippon Pharma, and has participated in advisory boards for Merck. LM has received consulting fees, honoraria, support for attending meetings and/or travel, and participated in Data Safety Monitoring Board or Advisory Boards for Bayer and Sanofi Aventis and has received support for attending meetings and/or travel and participated in Data Safety Monitoring Board or Advisory Boards for MSD. MJR has received grant funding from Allergan, Contrafect, Melinta, Merck, Paratek, Shionogi, T2 Biosystems, Tetraphase, consulting fees from Allergan, Merck, Shionogi, Spero, Ferring, and honoraria from Allergan, Abbvie, Shionogi. All other authors report no conflicts of interest.
Compliance with Ethics Guidelines
This study was approved by the Wayne State University institutional review board and waiver of informed consent was granted. The study was also approved by a human subject research committee at each study site when necessary and was conducted in accordance with the principles of the Declaration of Helsinki of 1964, and it's later amendments.
Prior Presentation
Parts of these data have been previously presented at the European Congress of Clinical Microbiology and Infectious Diseases in Vienna, Austria and IDWeek in San Francisco, CA, USA.
Data Availability
The datasets from this study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Zasowski, E.J., Trinh, T.D., Claeys, K.C. et al. International Validation of a Methicillin-Resistant Staphylococcus aureus Risk Assessment Tool for Skin and Soft Tissue Infections. Infect Dis Ther 11, 2253–2263 (2022). https://doi.org/10.1007/s40121-022-00712-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40121-022-00712-x