Abstract
Introduction
There is a need for data to evaluate hepatitis B antigenemia in newborns of mothers with hepatitis B virus (HBV) infection. This study aims to investigate this.
Methods
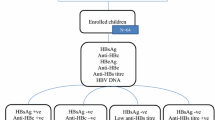
Newborns with positive serum hepatitis B surface antigen (HBsAg) and/or e antigen (HBeAg) were enrolled in the study.
Results
One hundred and one newborns from 98 HBV-infected mothers were included. Median maternal serum HBV DNA level was 23,200 IU/mL at delivery. Among the newborns, 48 were boys and 53 were girls. Mean birth weight was 3190.5 g. Twenty-one newborns had concurrent seropositive HBsAg and HBeAg, nine had seropositive HBsAg and seronegative HBeAg, and 71 had seronegative HBsAg and seropositive HBeAg. Eight newborns had detectable serum HBV DNA. In the follow-up, serum HBsAg and HBeAg in the newborns with undetectable HBV DNA became negative before 6 months of age. Two infants with detectable HBV DNA were diagnosed with immunoprophylaxis failure, one of whom developed active hepatitis at 3 months of age. Liver biopsy in this case showed significant interface hepatitis, fibrous septa formation, and expansion of portal areas with occasional bridging fibrosis.
Conclusions
Concurrent HBV viremia and antigenemia in newborns of HBV-infected mothers requires attention, while antigenemia without viremia is often transient.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Why carry out this study? |
Mother-to-child transmission is the major route of HBV infection. |
It is well known that active and passive immunoprophylaxis at birth does not exclude transmission of HBV infection. |
What was learned from the study? |
Concurrent HBV viremia and antigenemia in newborns of HBV-infected mothers requires attention, while antigenemia without viremia is often transient. |
Presentation of chronic hepatitis in histology, mixed genotpye infection and HBV DNA integration were found in a 3-month-old infant with immunoprophylaxis failure. |
Introduction
Hepatitis B virus (HBV) is a major public health threat since chronic infection can lead to liver cirrhosis and cancer [1,2,3]. Mother-to-child transmission is the major route of HBV infection in endemic areas [4]. A recent national investigation showed that 5.60 million pregnant women in China tested positive for hepatitis B surface antigen (HBsAg) between 2015 and 2020 [5]. Neonates born to HBsAg-positive mothers are at highest risk for development of chronic infection [6,7,8,9]. Despite passive–active immunoprophylaxis using hepatitis B vaccination with or without hepatitis B immunoglobulin (HBIg), up to 8–10% of newborns still acquire HBV infection [10].
Mother-to-child transmission of HBV may occur prenatally, during delivery, or early post partum. Chronic HBV infection in infancy can lead to one in four dying of HBV-related liver diseases in later life [11]. Given the consequences of mother-to-child transmission of HBV, close follow-up should be conducted in the newborns of HBV-infected mothers. Therefore, we performed the present clinical follow-up study to evaluate hepatitis B surface and/or e antigenemia in newborns of mothers with HBV infection.
Methods
Study Population
From January 2012 to December 2019, newborns (within 24 h after birth) who had positive serum HBsAg and/or hepatitis B e antigen (HBeAg) and had no obvious defects were enrolled in the multicenter observational cohort study. Their serological outcomes before hepatitis B vaccination and HBIg administration were provided as baseline parameters. Informed consent was obtained from the parents of each infant at admission. The study conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the ethics committees of each hospital.
Serological Assay
Serum HBsAg, anti-HBs, and HBeAg were determined using reagents from Roche Diagnostics (Roche Diagnostic GmbH, Mannheim, Germany). Serum HBV DNA was quantified by polymerase chain reaction (Roche COBAS AmpliPrep, lower limit of detection: 20 IU/mL).
Histological Evaluation
Liver biopsy specimens were prepared with hematoxylin and eosin staining, fiber staining, and immunohistochemical staining. Pathological evaluation was performed by an experienced hepatopathologist. Intrahepatic HBsAg and hepatitis B core antigen (HBcAg) were detected by immunohistochemistry and staining with brown particles was considered as positive expression.
Intrahepatic Covalently Closed Circular DNA (cccDNA) Quantification
Total viral DNA was extracted from formalin-fixed paraffin-embedded (FFPE) biopsy liver specimens using the QIAamp FFPE DNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Plasmid-safe ATP-dependent DNase enzyme (Epicentre, Madison, Wisconsin, USA) was used to digest HBV relaxed circular DNA, double-stranded linear DNA and single-stranded DNA. Rolling circle amplification was then conducted to selectively amplify circle DNA. Detailed procedure can be found in the reference paper [12].
HBV Genotype and Gene Integration Detection
To detect the integration of HBV DNA into human genome samples, targeted HBV DNA fragment capture sequencing was performed. DNA was extracted from liver tissues using GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany). Sequence capture probes according to eight types (A, B, C, D, E, F, G, and H) of HBV genome sequences were produced by MyGenostics (Beijing, China). Targeted HBV DNA fragment capture sequencing procedure was carried out according to a previous report [13]. Then, the high-throughput viral integration detection (HIVID) method was used for the detection of breakpoints [14]. After removal of low-quality and duplicate reads, sequenced datasets were compared against both HBV and human (hg19) reference sequences using Burrows–Wheeler aligner (BWA). Integration sites were determined through Clipping Reveals Structure (CREST) analysis. These integration sites were further confirmed by Sanger sequencing.
Whole-Exome Sequencing
Genomic DNA was extracted from peripheral blood. The detailed sequencing procedure can be found in our previous paper [15].
Statistical Analysis
Data analyses were performed using SAS 9.4 software (SAS Institute Inc, Cary, NC, USA). Continuous data were expressed as median (range) or mean ± standard deviation. Categorical data were expressed as the number of subjects or percentages. Group comparisons were performed using Fisher’s exact test. Tests were two-sided and a probability (P)-value of less than 0.05 was considered statistically significant.
Results
Baseline Characteristics
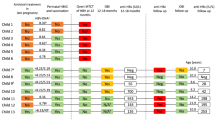
A total of 101 newborns were included in the study, including three pairs of twins. The babies were born from 98 HBV-infected mothers (mean age, 29.7 ± 4.2 years). All the mothers had seropositive HBsAg and 89 had seropositive HBeAg. Maternal serum median HBV DNA level was 23,200 IU/mL (range < 20–608,200,000 IU/mL) at delivery. Sixty-five mothers had received nucleoside analogue (NA) therapy in late pregnancy. Forty-two mothers delivered their babies through caesarean section. Table 1 shows the baseline characteristics of mothers at delivery.
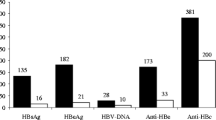
Among the newborns, 48 were boys and 53 were girls. Mean birth weight of these newborns was 3190.5 g. Twenty-one newborns had concurrent seropositive HBsAg and HBeAg, nine had seropositive HBsAg and seronegative HBeAg, and 71 had seronegative HBsAg and seropositive HBeAg. Three HBeAg-positive newborns were from HBeAg-negative mothers. Twenty-nine newborns had seropositive anti-HBs, of whom seven had seropositive HBsAg. Eight newborns had detectable serum HBV DNA, of whom two had negative serum HBsAg. Table 2 shows the baseline characteristics of newborns at birth. Six newborns with detectable HBV DNA were from mothers who did not receive NA therapy. There was a significant difference between the group of mothers who received NA therapy and mothers who did not receive NA therapy with respect to the distribution of newborns with detectable HBV DNA (P = 0.0165). All the newborns were administrated with hepatitis B vaccination and HBIg.
Findings in the Follow-Up
To date, included newborns have been followed for a median of 93 months (range 27–120 months). Serum HBsAg and HBeAg in the newborns with undetectable HBV DNA became negative before 6 months of age; however, three of them did not have positive serum anti-HBs (negativity, < 10 IU/L) on top of completion of the routine 3-dose vaccination. Among the newborns with detectable HBV DNA, six spontaneously cleared HBV DNA and HBsAg before 6 months of age, one developed active hepatitis at 3 months of age, and one had persistent infection with a high viral load in serum.
The infant who developed active hepatitis underwent further examinations. Her serum alanine transaminase (ALT), aspartate transaminase (AST), and HBV DNA were 112 U/L, 109 U/L, and 10,000,000 IU/mL, respectively. Coinfection with other hepatitis viruses (A, C, D, E), Epstein–Barr virus, cytomegalovirus, or human immunodeficiency virus was excluded. Whole-exome sequencing did not reveal clinically relevant variants. Liver biopsy showed significant interface hepatitis, fibrous septa formation, and expansion of portal areas with occasional bridging fibrosis (Fig. 1A, B). Immunohistochemical staining displayed intrahepatic positive HBsAg and HBcAg expression (Fig. 1C, D). Mixed HBV genotype infection was identified in her biopsy specimen, including genotypes A (4.13%), B (8.75%), and C (87.12%) (Fig. 2). The intrahepatic cccDNA quantification was 0.1767 copies/cell. Gene integration detection showed that there were 272 HBV DNA integration sites in the patient’s genome, of which 136 were intragenic.
Pathological presentations of a 3-month-old infant with hepatitis B. (A) significant interface hepatitis in hematoxylin–eosin staining biopsy liver tissue (× 200); (B) fibrous septa formation and expansion of portal areas with occasional bridging fibrosis in fiber staining (× 200); (C) intrahepatic positive HBsAg expression (brown particles) on immunohistochemical staining (× 400); (D) intrahepatic positive HBcAg expression (brown particles) on immunohistochemical staining (× 400)
Discussion
Current practice guidelines recommend that all pregnant women must be screened for HBV to identify the infection and further prevent vertical transmission [16]. NAs, such as lamivudine, telbivudine, and tenofovir, may be administered to inhibit viral replication in HBsAg-positive pregnant women with high HBV DNA levels. This prophylaxis significantly diminishes maternal HBV viremia at delivery, thereby reducing the risk of mother-to-child transmission [17]. In our study, newborns with detectable serum HBV DNA were mainly from the mothers who did not receive NA therapy, which indicated that appropriate prenatal prophylaxis had benefits in blocking mother-to-child transmission of HBV.
There has been controversy regarding newborns with antigenemia or viremia. Papaevangelou held the opinion that HBV viremia in newborns of HBsAg-positive mothers was a transient phenomenon that did not necessarily imply HBV infection transmission, whereas Alavian argued that transmission of HBV infection from HBsAg-positive mothers to infants needed more attention considering that several infants still acquired the infection on top of a combination of hepatitis B vaccination and HBIg [18, 19]. In the present cohort study, antigenemia among the newborns without viremia was absent in the follow-up; however, newborns with antigenemia and viremia did not all lose their HBsAg or HBeAg. As a result, as serum HBV DNA load was not known, antigenemia in newborns should not be underestimated. Additionally, serum anti-HB titers in the population are of concern. One study demonstrated that young children without seroprotective anti-HBs could be at risk of HBV infection through horizontal transmission [20]. For the three infants who had negative serum anti-HBs on top of completion of the routine three-dose vaccination in our study, a booster vaccination was administrated. However, to date, their anti-HB titers are still below 10 IU/L.
The diagnostic criteria of immunoprophylaxis failure are generally known as HBsAg positivity in infants older than 6 months after completion of a hepatitis B vaccination course [16]. In our study, immunoprophylaxis failure occurred in two infants. Although an infant developed active hepatitis at 3 months of age, this case can be diagnosed as immunoprophylaxis failure based on the biochemical, virological, and pathological outcomes. It is worth noting that the histological evaluation for this case showed overt chronic hepatitis identified by fibrous septa formation and expansion of portal areas with occasional bridging fibrosis. Additionally, many HBV DNA integration sites in the patient’s genome were identified. These findings are novel and have important clinical implications. As the first study that has focused on HBV DNA integration in pediatric HBV-infected liver tissues, the authors indicate that integration of HBV DNA to the host genome has rarely been confirmed at the early stage of chronic hepatitis in children [21]. With the development of detection methods, HBV DNA integration can be better explored [22]. Our study is the first to confirm numerous HBV DNA integration sites in a 3-month-old infant genome using this advanced detection method.
Many studies have demonstrated that different HBV genotypes have significant roles in determining the clinical prognosis of liver disease [23]. Several reports have shown that infection with mixtures of HBV genotypes is frequent, and coinfection with different HBV genotypes is associated with clinical disease severity [24, 25]. In our study, we found that the infant with active hepatitis was also infected with mixtures of HBV genotypes by high-throughput sequencing that provided more accurate information of the HBV genotype than PCR-based genotyping [26]. We inferred that the mixed HBV genotype infection might contribute to altered pathogenesis in this case.
Whether the integration of HBV DNA to the host genome in the infant patient plays a role in the disease progression remains unclear. This can be considered as a limitation of the current study and deserves further research.
Conclusions
Concurrent HBV viremia and antigenemia in newborns of HBV-infected mothers requires attention, while antigenemia without viremia is often transient. The findings in a 3-month-old infant with immunoprophylaxis failure may challenge current management of HBV infection.
References
Papatheodoridi M, Papatheodoridis G. New concepts regarding finite oral antiviral therapy for HBeAg-negative chronic hepatitis B. J Hepatol. 2021;75(6):1495–6.
Robbiani DF. Neutralizing hepatitis B. J Exp Med. 2020;217(10): e20201261.
Shi Y, Zheng M. Hepatitis B virus persistence and reactivation. BMJ. 2020;370: m2200.
Hui Z, Nayagam S, Chan P, et al. Progress towards elimination of mother-to-child transmission of hepatitis B virus infection in China: a modelling analysis. Bull World Health Organ. 2021;99(1):10–8.
Liu J, Wang X, Wang Q, et al. Hepatitis B virus infection among 90 million pregnant women in 2853 Chinese counties, 2015–2020: a national observational study. Lancet Reg Health West Pac. 2021;16: 100267.
Zhu S, Dong Y, Wang L, et al. Early initiation of antiviral therapy contributes to a rapid and significant loss of serum HBsAg in infantile-onset hepatitis B. J Hepatol. 2019;71(5):871–5.
Lu FT, Ni YH. Elimination of mother-to-infant transmission of hepatitis B virus: 35 years of experience. Pediatr Gastroenterol Hepatol Nutr. 2020;23(4):311–8.
Boucheron P, Lu Y, Yoshida K, et al. Accuracy of HBeAg to identify pregnant women at risk of transmitting hepatitis B virus to their neonates: a systematic review and meta-analysis. Lancet Infect Dis. 2021;21(1):85–96.
Veronese P, Dodi I, Esposito S, et al. Prevention of vertical transmission of hepatitis B virus infection. World J Gastroenterol. 2021;27(26):4182–93.
Shih YF, Liu CJ. Mother-to-infant transmission of hepatitis B virus: challenges and perspectives. Hepatol Int. 2017;11(6):481–4.
Doran Brubaker S, Ward JW, Hiebert L, et al. Developing an evidence base for the delivery of hepatitis B virus birth dose vaccination: an evidence map and critical appraisal of systematic reviews and guidelines. Clin Liver Dis (Hoboken). 2021;17(5):375–81.
Li W, Zhao J, Zou Z, et al. Analysis of hepatitis B virus intrahepatic covalently closed circular DNA and serum viral markers in treatment-naive patients with acute and chronic HBV infection. PLoS ONE. 2014;9(2): e89046.
Zhao LH, Liu X, Yan HX, et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. 2016;7:12992.
Li W, Zeng X, Lee NP, et al. HIVID: an efficient method to detect HBV integration using low coverage sequencing. Genomics. 2013;102(4):338–44.
Dong Y, Wang J, Zhu S, et al. Clinical profiles and diagnostic challenges in 1158 children with rare hepatobiliary disorders. Pediatr Res. 2021;89(1):238–45.
Sirilert S, Tongsong T. Hepatitis B virus infection in pregnancy: an update on evidence-based management. Obstet Gynecol Surv. 2020;75(9):557–65.
Kang B, Yi DY, Choe BH. Translational strategies to eliminate chronic hepatitis B in children: prophylaxis and management in East Asian countries. Front Pediatr. 2022;4(9): 809838. https://doi.org/10.3389/fped.2021.809838.
Papaevangelou V. Perinatal HBV viremia in newborns of HBsAg(+) mothers is a transient phenomenon that does not necessarily imply HBV infection transmission. J Clin Virol. 2012;54(2):202.
Alavian SM. Transmission of HBV infection from mothers HBsAg positive to infants need to more attention. J Clin Virol. 2012;54(2):201.
Pan XB, Yu J, Li HJ, et al. Young Chinese children without seroprotective hepatitis B surface antibody could be at risk of hepatitis B virus infection through horizontal transmission. J Viral Hepat. 2020;27(4):456–60.
Huang HP, Tsuei DJ, Wang KJ, et al. Differential integration rates of hepatitis B virus DNA in the liver of children with chronic hepatitis B virus infection and hepatocellular carcinoma. J Gastroenterol Hepatol. 2005;20(8):1206–14.
Zhao K, Liu A, Xia Y. Insights into hepatitis B virus DNA integration-55 years after virus discovery. Innovation (N Y). 2020;1(2): 100034.
Tanwar S, Dusheiko G. Is there any value to hepatitis B virus genotype analysis? Curr Gastroenterol Rep. 2012;14(1):37–46.
Liu CJ, Kao JH, Chen DS. Mixed hepatitis B virus genotype infections: the more, the worse? Hepatology. 2006;44(3):770.
Toan NL, le Song H, Kremsner PG, et al. Impact of the hepatitis B virus genotype and genotype mixtures on the course of liver disease in Vietnam. Hepatology. 2006;43(6):1375–84.
Xiao Y, Ni L, Cui Z, et al. Discrepant results of hepatitis B virus genotype determination by PCR and DNA sequencing. J Virol Methods. 2022;303: 114503.
Acknowledgements
We are grateful to Dr. Weijie Li for her great help in cccDNA quantification.
Funding
This study was supported by Beijing Natural Science Foundation (No. 7202193) and Beijing Municipal Science and Technology Commission (No. Z181100001718035). The journal’s Rapid Service Fees are being funded by the authors.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contributions
Pan Zhao designed the study. Jing Qi, Huijuan Liu, Ying Chen, Jiahui Fu, Huanwei Zheng, Pan Zhao and Limin Wang were responsible for data collection. Pan Zhao, Limin Wang, Jing Qi, Chunya Wang, Jing Chen and Ruifang Wang analysed the data. Pan Zhao and Jing Qi wrote the manuscript. Pan Zhao revised the manuscript. All authors reviewed and approved the final version of the manuscript.
Disclosures
All names authors confirm that they have nothing to disclose.
Compliance with Ethics Guidelines
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the ethics committees of each hospital.
Data Availability
The data used for this study are not freely available because of human participants. Interested researchers can contact the corresponding authors through email for more detailed information.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Qi, J., Liu, H., Wang, L. et al. Follow-Up of Newborns with Hepatitis B Antigenemia. Infect Dis Ther 11, 2233–2240 (2022). https://doi.org/10.1007/s40121-022-00704-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40121-022-00704-x