Abstract
Introduction
The causal association between the gut microbiome and the risk of intracranial aneurysm (IA), subarachnoid hemorrhage (SAH), and unruptured aneurysm (uIA) is unclear.
Methods
The single nucleotide polymorphisms concerning gut microbiome were retrieved from the gene-wide association study (GWAS) of the MiBioGen consortium. The summary-level datasets of IA and SAH were obtained from the GWAS meta-analysis of the International Stroke Genetics Consortium (ISGC). Inverse variance weighting (IVW) was utilized as the primary method, complemented with sensitivity analyses for pleiotropy and increasing robustness.
Results
Five, seven, and six bacterial traits were found to have a causal effect on IA, SAH, and uIA, respectively (IVW, all P < 0.05). Family.Porphyromonadaceae and genus.Bilophila were common protective bacterial features for both SAH and uIA. The heterogeneity and pleiotropy analyses confirmed the robustness of IVW results.
Conclusion
Our study demonstrates that gut microbiomes may exert therapeutic effects on IA, uIA, and SAH, providing clinical implications for the development of novel biomarkers and therapeutic targets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Why carry out this study? |
The causal association between the gut microbiome and the risk of intracranial aneurysm (IA), unruptured aneurysm (uIA), and subarachnoid hemorrhage (SAH) is unclear. |
We have used a two-sample Mendelian randomization approach to investigate the causal effects of gut microbiome on IA, uIA, and SAH in this study. We also conducted the bidirectional MR analysis to explore the association between gut microbiome and SAH. |
What was learned from the study? |
Our study shows novel genetic evidence that gut microbiomes may exert therapeutic effects on the development of IA, uIA, and SAH, providing clinical implications for the development of novel biomarkers and therapeutic targets. |
No causal effects of SAH on gut microbiome were identified in reverse MR analysis. |
Introduction
Intracranial aneurysm (IA) is characterized by balloon-shaped dilatation and has a prevalence of about 3.2% in the general population [1]. The rupture of IA leading to subarachnoid hemorrhage (SAH) is a life-threatening type of stroke. Approximately one-third of patients with SAH die within 24 h [2]. Despite the results of recent genetic studies suggesting the involvement of specific genes CDKN2A, SOX17, and ADAMTS15 in SAH, the identified variants explain only a small proportion of population-based heritability [3].
Mounting evidence has implied that gut microbiota play an important role in host metabolism and immune homeostasis. The effect of gut microbiota on cardiovascular diseases, including hypertension [4] and heart failure [5], has recently explored. These studies demonstrated that metabolites from gut microbiota can be absorbed into the circulation and exert effects on tissue injury and/or repair [5, 6]. Furthermore, the depletion of gut microbiota using antibiotics can significantly decrease the incidence of IA [7], indicating that inflammation may contribute to the pathophysiology of the aneurysm wall [8]. However, gut microbiota produce a functional complex of biomolecules, and it remains unknown whether one or multiple bacterial traits are involved in the development of IA and SAH. Clinical studies and preclinical experiments have potential shortcomings such as limited sample size and retrospective design, impeding our understanding of these complex diseases.
Mendelian randomization (MR) is a robust and effective method using genetic variants (single nucleotide polymorphisms, SNPs) to overcome the aforementioned shortcomings [9]. A random assortment of SNPs are involved in the formation of a zygote during gestation [10], and the random assortment of interventions is similar to the assignment in a randomized clinical trial [10, 11]. Therefore, MR study can yield the causal association between the exposure (gut microbiome) and outcomes (IA, SAH, and unruptured IA [uIA]). Furthermore, MR analyses may have enough power to detect a causal effect. At present, the causal association between gut microbiota and IA, SAH, and uIA is lacking. To reveal the effect of the gut microbiome on IA, SAH, and uIA, we performed a two-sample MR analysis to explore their potential causal links.
Methods
Data Sources of gut microbiome
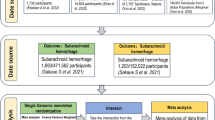
The summary-level datasets for gut microbiome were retrieved from the gene-wide association study (GWAS) of the MiBioGen consortium [12], including a total of 18,340 samples of 16S rRNA gene sequencing data from 24 population-based cohorts. A total of 211 gut microbiomes from genus to phylum level were identified. All datasets were analyzed by three 16S rRNA regions and rarefied to 10,000 reads for rarefaction reproducibility, with 131 genera, 16 classes, 35 families, 20 orders, and 9 phyla being identified. Detailed information on the gut microbiome was described in the original article [12].
Data Sources of IA, aSAH, and uIA
The summary-level datasets regarding IA (7495 cases and 71,934 controls), SAH (5140 cases and 71,934 controls), and uIA (2070 cases and 71,934 controls) were extracted from the GWAS meta-analysis of the International Stroke Genetics Consortium [13]. The IA includes unruptured IA and ruptured IA. Summary-level datasets of IA, SAH, uIA, and gut microbiome were extracted from distinct GWAS projects.
Compliance with Ethics Guidelines
All summary-level datasets in our study were extracted from de-identified public data/studies. All studies were approved by the ethics committee previously and informed consent was obtained from all participants in original studies [12, 13]. Ethical approval is thus exempted from our study because we only used the summary-level data. The Strengthening the Reporting of Observational Studies in Epidemiology using Mendelian Randomization (STROBE-MR) checklist is provided in the supplementary material. The study was in accordance with the Declaration of Helsinki.
Genetic Instrument Selection
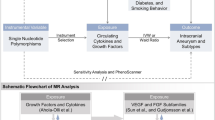
The genetic instruments associated with bacterial traits were selected at a genome-wide significance level (P < 1 × 10−5). The independence of SNPs was evaluated with the threshold of an r2 < 0.01 and clumping window (10,000 kb). The instrument variables (IVs) obtained are shown in Table S2 in the supplementary material. Furthermore, MR Pleiotropy RESidual Sum and Outlier (MR-PRESSO) approach was utilized to explore significant SNPs accounting for possible pleiotropy [14], and the outlier SNPs were removed. The results of F statistics = (Bets/Se)2 represent the strength of MR, and the value of F statistics > 10 is an indicator of statistical robustness [15]. In this formula, beta is the correlation coefficient between SNPs and traits (bacterial trait and IA). All values of F statistics exceed 10 in this MR study. In addition, we also explored the causal effects of SAH on gut microbiome in reverse MR analysis.
Main Statistical Analyses
The random effects inverse variance weighting (re-IVW) approach is the primary method to explore the causal associations in the MR study since this method can provide a robust causal estimate in the absence of directional pleiotropy (no violation of the independence assumption). P < 0.05 represents statistical significance. All analyses were conducted using “TwoSampleMR”, “mr.raps”, “MRPRESSO”, “frostplot”, and “ggplot2” in the R software (version 4.2.0, The R Foundation, Vienna, Austria).
Sensitivity Analyses
A variety of methods including MR-Egger, weight median, maximum likelihood, MR robust adjusted profile score (MR-RAPS), and MR-PRESSO were performed for sensitivity analyses. The potential horizontal pleiotropy was explored using MR-Egger and MR-PRESSO [14, 16]. Weighted median-based MR analysis was also used to evaluate the causal effect, assuming that at least half of the IVs are valid [17]. Cochran’s Q statistic was applied to explore the heterogeneity among variant-specific estimates. Similar to IVW, the maximum likelihood method can be used when heterogeneity and horizontal pleiotropy do not exist. Moreover, compared to IVW, the standard errors are smaller in maximum likelihood analysis [18]. MR-RAPS analysis can yield robust inference to systematic and idiosyncratic pleiotropy when many weak instruments exist [19]. In addition, leave-one-out analysis was performed to verify the robustness of the conclusion.
Results
Genetic Instrument Variables for Gut Microbiome
A total of 211 bacterial traits including five biological levels (phylum, class, order, family, and genus) were collected in our study. SNPs used as IVs for each bacterial trait are summarized in Table S2. Fifteen bacterial traits were removed as a result of unknown traits. Furthermore, genus.LachnospiraceaeND3007 group was deleted because no SNP is harmonized and the IVW results cannot be obtained in MR analysis for uIA and SAH. Collectively, 196, 195, and 195 bacterial traits were included in MR analyses for IA, SAH, and uIA, respectively. Positive MR results of causal links between gut microbiome and IA, SAH, and uIA are shown in Table 1.
Casual Effect of the Genetically Predicted Gut Microbiome on IA
The causal effects of 196 bacterial taxa on IA risk are shown in Fig. 1a. In the condition of IA, three bacterial traits causally related to the risk of IA were identified. Specifically, Genus.CandidatusSoleaferrea was shown to decrease the risk of IA (odds ratio [OR] 0.85, 95% confidence interval [CI] 0.77–0.95, P = 0.0041, Fig. 2, Table S3 in the supplementary material), while genus.Holdemania and genus.Olsenella were associated with an increased risk of IA (genus.Holdemania, OR 1.15, 95% CI 1.01–1.29, P = 0.0289; genus.Olsenella, OR 1.10, 95% CI 1.02–1.20, P = 0.0163, Fig. 2, Fig. S1A in the supplementary material). A similar trend was observed in the RAPS and maximum likelihood analyses (Fig. 2). Leave-one-out analyses showed no significant SNPs (Fig. S1B). The results of MR-Egger and MR-PRESSO analyses demonstrated no signs of pleiotropy (Table 2). Moreover, the results of Cochran’s Q test demonstrated no signs of heterogeneity (Table 2), as shown in the funnel plot (Fig. S1C). The Frost plot visualizing the effect estimate of each SNP for each bacterial trait on IA is displayed in Fig. S1D.
Causal effect of the gut microbiome on IA, uIA, and SAH based on MR analyses. From outside to inside, the P values of IVW, MR Egger, WMe, WMo, and SM are represented, respectively. The GM taxa name represented by each ID can be found in Table S1. a MR results of IA. b MR results of SAH. c MR results of uIA. IVW inverse variance weighted, WMe weighted median, WMo weighted mode, SM simple mode
Casual Effect of Genetically Predicted Gut Microbiome on SAH
The causal effects of 195 bacterial taxa on the risk of SAH are shown in Fig. 1b. Causal effects of genetically predicted seven bacterial traits on the risk of SAH were identified. Among these traits, six bacterial features were causally related to a decreased risk of SAH (class.Lentisphaeria, OR 0.79, 95% CI 0.62–0.98, P = 0.0472; family.Porphyromonadaceae, OR 0.66, 95% CI 0.47–0.95, P = 0.0245; genus.Bilophila, OR 0.68, 95% CI 0.50–0.93, P = 0.0167; genus.Fusicatenibacter, OR 0.63, 95% CI 0.46–0.88 P = 0.0058; genus.Ruminococcus1, OR 0.48, 95% CI 0.30–0.78, P = 0.0031; order.Victivallales, OR 0.79, 95% CI 0.62–0.99, P = 0.0472, Fig. 3, Table S4 in the supplementary material). In contrast, family.Streptococcaceae was positively related to an increased risk of SAH (OR 1.31, 95% CI 1.01–1.71, P = 0.0486, Fig. 3). The casual association between the gut microbiome and SAH was still noted in the RAPS and maximum likelihood analyses (Fig. 3). The leave-one-out analysis did not identify any significant outliers after removing each SNP (Fig. S2A in the supplementary material). In addition, no signs of pleiotropy were detected in MR-Egger and MR-PRESSO analyses (Fig. S2B, Table 2). Cochran’s Q test suggested no evidence of heterogeneity (Table 2), and the MR results in the funnel plot were symmetrical (Fig. S2C). The results of the MR analyses for each SNP in seven bacterial traits on SAH are shown in Fig. S2D.
Casual Effect of the Genetically Predicted Gut Microbiome on uIA
The causal effects of 195 bacterial taxa on uIA risk are shown in Fig. 1c. For uIA, our results demonstrated that six bacterial features were causally related to uIA (Table 3). Two of them decreased the risk of uIA (family.Porphyromonadaceae, OR 0.49, 95% CI 0.25–0.96, P = 0.0381; genus.Bilophila, OR 0.54, 95% CI 0.31–0.94, P = 0.0285; Table 3; Fig. S3A, Table S5 in the supplementary material), while four traits increased the risk of uIA (family.Oxalobacteraceae, OR 1.34, 95% CI 1.01–1.76, P = 0.0404; genus.Adlercreutzia, OR 1.73, 95% CI 1.09–2.75, P = 0.0191; genus.Intestinimonas, OR 1.44, 95% CI 1.02–2.03, P = 0.0364; genus.Victivallis, OR 1.38, 95% CI 1.01–1.88, P = 0.0437; Table 3, Fig. S3A). Leave-one-out test revealed that the causal association was still noted after leaving out any single SNP (Fig. S3B). In Cochran’s Q test, no signs of heterogeneity were detected (Fig. S3C, Table 2). Furthermore, no evidence of pleiotropy was observed in MR-Egger and MR-PRESSO analyses (Table 2). The forest plot visualizing the effect estimate of each SNP on uIA is displayed in Fig. S3D.
Casual Effect of the Genetically Predicted IA, SAH, uIA on Gut Microbiome
The IVs for SAH used in reverse MR analysis are shown in Table S6 in the supplementary material. No causal effects of SAH on gut microbiome were identified in the reverse MR analysis (Fig. S4 in the supplementary material).
Discussion
In this MR study, we identified the causal effect of bacterial entities in the gut microbiome on the risk of IA, SAH, and uIA. For IA, our results demonstrated that genetically predicted genus.CandidatusSoleaferrea is a protective element, while genus.Holdemania and genus.Olsenella may increase the risk. For SAH and uIA, we found that both familiy.Porphyromonadaceae and genus.Bilophila are protective factors. Genus.Fusicatenibacter and genus.Ruminococcus1 may decrease the risk of SAH. Regarding uIA, family.Oxalobacteraceae, genus.Adlercreutzia, genus.Intestinimonas, and genus.Victivallis may increase the risk of uIA. Our results may provide novel clues to illustrate the effect of specific bacterial features on the development of IA and the occurrence of SAH.
The development of IA is associated with an alteration of gut microbiota. Li and colleagues found that a total of 145 genera were differentially enriched between the patients with uIA and the controls [8]. The abundance of the genus.CandidatusSoleaferrea and family.Porphyromonadaceae was overrepresented in patients with uIA compared with controls [8]. However, they did not illustrate the role of specific bacterial features. Our results showed that the abundance of both bacterial features is highly increased in uIA. We also found that family.Porphyromonadaceae and genus.CandidatusSoleaferrea may exert protective effects in the development of IA, while several bacterial traits may increase the risk on the development of IA. The occurrence of SAH is closely associated with dysbiosis of the gut microbiome. The changes of gut microbiome profiles (Campylobacter and Campylobacter ureolyticus) are associated with the rupture of IA and occurrence of SAH [20]. In our MR study, the increased abundance of family.Porphyromonadaceae may play a protective role in the occurrence of SAH. Another five bacterial features may have similar effects in the condition of SAH.
Inflammation contributes to the development of IA and the occurrence of SAH [7, 20]. Porphyromonadaceae is capable of producing short-chain fatty acids (SCFAs), which have potent anti-inflammatory effects on immune cell functions. Specifically, SCFAs can exert immunomodulating effects by prompting cellular differentiation of CD25+ Foxp3+ Treg cells [21, 22]. Beneficial effects of SCFAs have been noted in several diseases, including colitis and ischemia–reperfusion kidney injury [23]. The protective role of family.Porphyromonadaceae in IA and SAH can be explained by anti-inflammatory effects of SCFAs. In addition, SCFAs may cause blood pressure reduction through renin release and vasomotor function [24], serving as a major risk factor for IA and SAH. The genus.CandidatusSoleaferrea exerted anti-inflammatory effects through secreting metabolites and intestinal homeostasis protection properties [25]. By contrast, detrimental effects of family.Streptococcaceae may be achieved through risk factors including obesity and hypertension. For example, the increased abundance of family.Streptococcaceae is associated with BMI and waist circumference [26], playing an important in IA and SAH. The family.Streptococcaceae may also contribute to higher blood pressure [27, 28].
As a non-invasive approach, the gut microbiome test may be performed to evaluate the risk of IA and SAH based on specific abundant species in the future, especially for individuals with high-risk factors including hypertension and obesity. Potential identification of bacterial traits may also provide valuable clues for therapeutic approaches. A recent study has shown that the depletion of gut microbiota decreased the incidence of mice IA [7], with the Hungatella hathewayi exerting a causal role in the formation and rupture of intracranial aneurysms [25]. In our MR analyses, a variety of bacterial traits were identified, and the combined benefits of bacterial features can be achieved through fecal transplantation.
Our study has several limitations. Firstly, the threshold filtering of the IVs has a relatively low significance level of P < 1 × 10−5. Secondly, GWAS summary-level data from European participants may limit the generalizability of our findings. Thirdly, we did not conduct a strict multiple correction test considering this method so potential bacterial taxa may have been overlooked. Furthermore, a large sample size is required to explore the link between gut microbiome and IA, SAH, and uIA as a result of the small sample size of gut microbiome. Finally, individual association could not be performed because of lack of individual data. Future studies are needed to verify our conclusions in large sample sizes and different ethnicity and to clarify potential mechanisms.
Conclusion
This MR study identifies causal links between the gut microbiome and the development of IA, uIA, and the occurrence of SAH. Gut microbiome composition may serve as a promising biomarker and therapeutic target for IA, uIA, and SAH.
References
Vlak MH, Algra A, Brandenburg R, Rinkel GJ. Prevalence of unruptured intracranial aneurysms, with emphasis on sex, age, comorbidity, country, and time period: a systematic review and meta-analysis. Lancet Neurol. 2011;10:626–36.
Nieuwkamp DJ, Setz LE, Algra A, Linn FH, de Rooij NK, Rinkel GJ. Changes in case fatality of aneurysmal subarachnoid haemorrhage over time, according to age, sex, and region: a meta-analysis. Lancet Neurol. 2009;8:635–42.
Korja M, Silventoinen K, McCarron P, et al. Genetic epidemiology of spontaneous subarachnoid hemorrhage: Nordic twin study. Stroke. 2010;41:2458–62.
Li J, Zhao F, Wang Y, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:14.
Cui X, Ye L, Li J, et al. Metagenomic and metabolomic analyses unveil dysbiosis of gut microbiota in chronic heart failure patients. Sci Rep. 2018;8:635.
Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63.
Shikata F, Shimada K, Sato H, et al. Potential influences of gut microbiota on the formation of intracranial aneurysm. Hypertension. 2019;73:491–6.
Li H, Xu H, Li Y, et al. Alterations of gut microbiota contribute to the progression of unruptured intracranial aneurysms. Nat Commun. 2020;11:3218.
Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318:1925–6.
Smith GD, Ebrahim S. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22.
Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65.
Kurilshikov A, Medina-Gomez C, Bacigalupe R, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021;53:156–65.
Bakker MK, van der Spek RAA, van Rheenen W, et al. Genome-wide association study of intracranial aneurysms identifies 17 risk loci and genetic overlap with clinical risk factors. Nat Genet. 2020;52:1303–13.
Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–8.
Chen L, Yang H, Li H, He C, Yang L, Lv G. Insights into modifiable risk factors of cholelithiasis: a Mendelian randomization study. Hepatology. 2022;75:785–96.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25.
Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14.
Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol. 2013;178:1177–84.
Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. 2020. https://arxiv.org/abs/1801.09652.
Kawabata S, Takagaki M, Nakamura H, et al. Dysbiosis of gut microbiome is associated with rupture of cerebral aneurysms. Stroke. 2022;53:895–903.
Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73.
Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–5.
Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–45.
Marques FZ, Nelson E, Chu PY, et al. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation. 2017;135:964–77.
Cai J, Zhou L, Song X, et al. Alteration of intestinal microbiota in 3-deoxyglucosone-induced prediabetic rats. Biomed Res Int. 2020;2020:8406846.
Verhaar BJH, Collard D, Prodan A, et al. Associations between gut microbiota, faecal short-chain fatty acids, and blood pressure across ethnic groups: the HELIUS study. Eur Heart J. 2020;41:4259–67.
Kim S, Goel R, Kumar A, et al. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin Sci (Lond). 2018;132:701–18.
Yan Q, Gu Y, Li X, et al. Alterations of the gut microbiome in hypertension. Front Cell Infect Microbiol. 2017;7:381.
Acknowledgements
We express gratitude to participants, researchers, and staff associated with the previous GWAS studies from which we used data for this MR study.
Funding
This study is supported by the National Natural Science Foundation of China (81701292). The Rapid Service Fee was funded by the authors.
Medical Writing and Editorial Assistance
The authors did not use medical writing or editorial support for this paper.
Author Contributions
Mei He and Wenjing Wang analyzed and interpreted the data and wrote the manuscript; Qiang He, Heling Dai, and Jinming Han analyzed the data; Jinming Han and Wenyao Cui designed the study and reviewed the manuscript.
Prior Presentation
This manuscript is based on work that has been previously presented/published by Kurilshikov A et al. and Bakker MK e al in Nature Genetics, respectively [12, 13].
Disclosures
All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Compliance with Ethics Statement
All summary-level datasets in our study were extracted from de-identified public data/studies. All studies were approved by the ethics committee previously and informed consent was obtained from all participants in original studies [12, 13]. Ethical approval is thus exempted from our study because we only used the summary-level data. The Strengthening the Reporting of Observational Studies in Epidemiology using Mendelian Randomization (STROBE-MR) checklist is provided in the supplementary material. The study was in accordance with the Declaration of Helsinki.
Data Availability
All the data in our MR study are publicly accessible (https://gwas.mrcieu.ac.uk/).
Author information
Authors and Affiliations
Corresponding authors
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
He, M., Wang, W., He, Q. et al. Genetic Causal Association Between the Gut Microbiome and Intracranial Aneurysm and Subarachnoid Hemorrhage: A Two-Sample Mendelian Randomization Study. Neurol Ther 12, 1695–1707 (2023). https://doi.org/10.1007/s40120-023-00525-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40120-023-00525-1