Abstract
Background
Since 2010, 27 mixed-treatment comparisons (MTCs) of disease-modifying therapies (DMTs) for multiple sclerosis have been published. However, there has been continued evolution in the field of MTCs. Additionally, limitations in methodological approach and reporting transparency, even in the most recent publications, makes interpretation and comparison of existing studies difficult.
Objectives
The objectives of this study are twofold: (1) to estimate the efficacy and safety of DMTs at European Commission-approved doses compared with placebo in adults with relapsing–remitting multiple sclerosis (RRMS) using MTC, and (2) to identify and address methodological challenges when performing MTC in RRMS, thereby creating a baseline for comparisons with future treatments.
Methods
Searches were completed in 14 databases, including MEDLINE, Embase, CENTRAL, CDSR and DARE, from inception to June 2018 to identify published or unpublished prospective, randomised controlled trials of all European Union-approved DMTs or DMTs expected to be approved in the near future in RRMS or rapidly-evolving severe RRMS. No language or date restrictions were applied. Studies were included in the MTC if they were judged to have sufficiently similar characteristics, based on the following: patient age; proportion of male participants; Expanded Disability Status Scale (EDSS) score; duration of disease; number of relapses prior to enrolment and proportion of previously treated patients. Background information from the included studies, as well as effect size and confidence intervals (where relevant) of defined outcomes were extracted. Reporting of the MTC was consistent with the International Society for Pharmacoeconomics and Outcomes Research (ISPOR) and Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) network meta-analysis guidelines.
Results
In total, 33 studies were included in the MTC. Annualised relapse rate (ARR 28 trials) was significantly reduced in all treatments compared with placebo. Alemtuzumab had the highest probability (63%) of being the most effective treatment in terms of ARR compared with placebo (rate ratio [RR] 0.28, 95% credible interval [CrI] 0.21–0.38), followed by natalizumab (30% probability; RR 0.32, 95% CrI 0.23–0.43). The risk of 3- and 6-month confirmed disability progression (CDP3M, 13 trials; CDP6M, 14 trials) were similar; CDP6M was significantly reduced for alemtuzumab (hazard ratio [HR] 0.365; 95% CrI 0.165–0.725), ocrelizumab (HR 0.405, 95% CrI 0.188–0.853) and natalizumab (HR 0.459, 95% CrI 0.252–0.840) relative to placebo. There were no significant differences in the odds of serious adverse events (SAEs, 6 trials) between any treatment and placebo. The results of the MTC were limited by the lack of studies reporting direct comparisons between the included treatments and by heterogeneous reporting of key outcome data.
Conclusions
Meta-analyses confirmed the benefit of all DMTs in terms of relapse rate compared with placebo with a comparable rate of SAEs for the DMTs that could be included in the network. The rigor and transparency of reporting in this study provide a benchmark for comparisons with future new agents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
A number of disease-modifying therapies (DMTs) are approved for the treatment of multiple sclerosis, but direct comparative data do not exist for each individually with all other alternative options. |
The results of this mixed treatment comparison (MTC) confirmed that all DMTs included were beneficial in terms of relapse rate when compared with placebo, with a comparable rate of serious adverse events. |
The convergence of evidence from different sources may help to provide more precision to conclusions when comparing across the treatment arena. |
The rigorous and transparent approach taken allows the results of this MTC to be held as a benchmark for comparisons with future new agents. |
Digital Features
This article is published with digital features to facilitate understanding of the article. You can access the digital features on the article’s associated Figshare page. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.12912446.
Introduction
While the use of mixed treatment comparison (MTC) has become increasingly relevant in healthcare decision-making, this field continues to evolve and mature. Interpretation of MTC results can be complex [1,2,3] as findings for the same research question can vary depending on how a systematic review and a meta-analysis were performed or reported. Consequently, there is a growing interest in resolving the issues surrounding clinical MTC with best-practice recommendations and reporting guidelines [2, 4,5,6,7].
Multiple sclerosis (MS) is a chronic autoimmune-mediated inflammatory disease of the central nervous system that affects an estimated 2.3 million people worldwide [8, 9]. Ongoing demyelination of central nerve fibres results in patients become progressively more disabled over time [8]. Approximately 85% of patients diagnosed with MS have a relapsing–remitting disease type (RRMS) [9] and, therefore, can be considered for disease-modifying therapies (DMTs) [10]. Treatment with DMTs aims not to cure RRMS but instead reduce the rate of MS relapses and ultimately delay the progression of disability [10]. A number of DMTs are already approved for use in the European Union (EU; 14 at the time of analysis), but direct comparative data do not exist for each individual DMT with all other alternative options.
The diagnosis of MS has evolved over time from criteria initially described by Poser [11] to subsequent updates of McDonald criteria [12,13,14]. This development combined with temporal changes to definitions of outcome measures [15] make meeting the assumptions of similarity, homogeneity and transitivity, which are critical to optimum MTC design, challenging in MS [2, 16]. While undertaking an analysis of methodological approaches, Sormani et al. 2020 [17] identified 27 MTCs of treatments for MS reported since 2010, and also identified substantial differences between the reported studies in terms of the inclusion criteria, treatments assessed and analysis methods which made it impossible to directly compare findings. In general, the estimated treatment effects relative to placebo were in the same direction for those reported by multiple studies, and this trend was consistent across outcomes and populations. However, the magnitude of the effects and the associated level of uncertainty for each finding varied as a result of the differences between MTCs [17]. Even in one of the most recently published MTCs, the methodological approach employed was compromised by combining different time points for a shared endpoint measure (12-and 24-month annualised relapse rate [ARR]) from different trials, the inclusion of off-label treatments or doses and the use of less stringent endpoints for measuring disability (3- rather than 6-month confirmed disability progression) [18]. Given this, our aim was to undertake the most rigorous MTC of DMTs in RRMS at European approved doses, thereby providing a benchmark for future MTCs in this disease area, and avoiding some the challenges in the interpretation of previous RRMS MTCs.
The objective of this study was to conduct a systematic literature review and MTC to estimate the clinical efficacy, in terms of relapse rate and progression of disability over time, and safety, in terms of risk of serious adverse events (SAEs), of DMTs at European Commission-approved doses compared with placebo in adults with RRMS.
Methods
The systematic review methods adhered to published methods [19,20,21]. This study was reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) network meta-analysis [4] (Electronic Supplementary Material [ESM] Appendix 1) and International Society for Pharmacoeconomics and Outcomes Research (ISPOR) guidelines [22].
This study included prospective randomised controlled trials (RCTs) of adults (aged ≥ 18 years) with a confirmed diagnosis of RRMS (≥ 85% of the study population) and treated with dimethyl fumarate (DMF), interferon (IFN) beta-1a, pegylated IFN (IFN) beta-1a, IFN beta-1b, natalizumab, glatiramer acetate (GA), fingolimod, teriflunomide, alemtuzumab, ocrelizumab, cladribine or placebo. This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
The key outcomes of interest for the MTC were ARR, 3- and 6-month confirmed disability progression (CDP3M and CDP6M, respectively) and SAEs.
MEDLINE, MEDLINE In-Process, MEDLINE Daily Update, MEDLINE Epub Ahead of Print, PubMed, Embase, Cochrane Central Register of Controlled Trials (CENTRAL), Science Citation Index (SCI), National Institutes of Health (NIH) ClinicalTrials.gov, World Health Organization (WHO), International Clinical Trials Registry Platform (ICTRP), PharmNetBund, EU Clinical Trials Register (EUCTR), International Standard Randomised Controlled Trial Number (ISRCTN) Registry, electronic medicines compendium (eMC) and European Medicines Agency (EMA) register were searched for relevant studies from database inception to June 2018 without language or publication limits. The MEDLINE search strategy is shown in ESM Appendix 2. The reference lists of included articles were checked for additional relevant studies.
Two reviewers independently screened articles for inclusion, assessed study quality and performed data extraction. For each study, the background information (year of publication, other related publications, country, funding, study aim and treatment type) were extracted where available. Other specific data extracted were sample size, location/setting, methods employed (e.g. randomisation and allocation concealment, blinding), patient baseline characteristics (e.g. age, diagnosis, comorbidities, previous and concomitant treatments), interventions/study arms compared (description of interventions and comparators), outcomes assessed (e.g. definition of outcome, when assessed, who assessed, methods used to assess outcome[s]), results (e.g. numbers, percentages and effect sizes with confidence intervals [CIs; where relevant]) and follow-up time. Notably, for the outcomes of interest, ARR per arm was extracted as the total number of relapses/total number of patient-years of follow-up. For time-to-event outcomes (e.g. disability progression) the hazard ratio (HR) with 95% CI was extracted, where possible. SAE data were extracted according to the definition of each individual study, excluding MS relapse. The methodological quality of each study was assessed using the Cochrane risk-of-bias tool for RCTs [23]. Discrepancies at all stages of the review were resolved through discussion or consultation with a third reviewer.
Networks were created for each of ARR, CDP3M, CDP6M and SAE. ARR was analysed using the rate ratio (RR) at any time point ≥ 12 months. This cutoff was considered to be sufficient as ARR is essentially a value determined over 12 months. Less than 12 months was considered too short an interval to show a clinical effect. CDP3M or CPD6M were analysed using the HR for time-to-disability progression at 24 months as the effect estimate. SAE used the odds ratio (OR) at the 24-month follow-up as the effect estimate. The studies eligible for inclusion in the network were assessed for similarity based on the following characteristics: diagnosis, diagnostic criteria, age, gender, Expanded Disability Status Scale (EDSS) range, duration of disease, number of relapses prior to enrolment, previously treated patients, EU licensed doses only and follow-up time.
‘Head-to-head’ comparisons of treatments were performed in line with the Cochrane Handbook for Systematic Reviews of Interventions [20]. Forest plots of effect sizes, showing the results of individual studies, were prepared using the meta-package [24] in R software (R Foundation for Statistical Computing, Vienna, Austria) [25]. Where more than one study reported the same outcome measure for clinically similar populations, pooled effect estimates and 95% CIs were calculated using random-effects models. For pooled analyses of relative treatment effects (e.g. RR, HR), study weights were calculated using the generic inverse variance method. For pooled analyses of binary data based on the number of participants with an event and the total number of participants, study weights were calculated using the Mantel–Haenszel method. Assessment of publication bias was not possible due to a lack of sufficient studies. Heterogeneity was assessed using the I2 statistic.
All indirect comparisons and MTC methods used in this report are consistent with ISPOR task force recommendations for the conduct of direct and indirect meta-analyses [6, 7]. MTC was performed using a Bayesian approach using the gemtc package [26]. A burn-in of 50,000 simulations was used, followed by a further run of 50,000 simulations to obtain parameter estimates. Model convergence was assessed using the Brooks–Gelman–Rubin statistic [27]. Random-effects models were used. Model fit was assessed using residual deviance and the deviance information criterion. A frequency table was constructed from these rankings and normalised by the number of iterations to give the rank probabilities [25]. Sensitivity analyses were performed for fixed-effects models, the definition of SAEs and follow-up data.
In many cases, the data required as inputs to standard MTC models were not reported by the included studies. The missing values were calculated from the available data using standard methods [20, 28,29,30]. When missing values could not be estimated from the available data they were not included in the analysis.
Network diagrams for each outcome were assessed for the presence of loops where inconsistency may occur. For those networks with the potential for inconsistency, we used the node splitting method to check for evidence of inconsistency [3]. An inconsistency factor of exactly 1 would indicate that the indirect and direct estimates of the treatment comparison were exactly equal (≥ 2 studies per comparison were required). To further confirm the results of the MTC analysis, pairwise meta-analysis was compared to the relevant analyses.
Results
Systematic Review
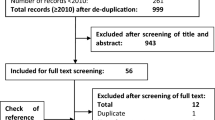
The search strategies identified 19,577 unique citations (Fig. 1). After title and abstract screening, 1366 articles met the criteria for full-text review, of which 537 were excluded. Fifty-one studies (779 publications) were included in the review; references can be found in ESM Appendix 3.
PRISMA diagram to illustrate the screening of studies from searches to inclusion. aNot available through any online resources or via the British Library. EPAR European public assessment report, PRISMA Preferred Reporting Items for Systematic Reviews and Meta-Analyses, SmPC summary of product characteristics
Creating and Defining the Network
The similarity of the study and patient characteristics were analysed to allow network definition. A summary of the inclusion criteria for all included studies is shown in ESM Appendix 4a. The oldest trial was published in 1987 [31], while over half of the included trials (28 of 51) were published since 2010. MS diagnosis was mainly based on the McDonald 2005 criteria [13] but there was some variation across trials. Most studies (84%) included over 100 patients, but seven included less [31,32,33,34,35,36,37]. Most (88%) had a cutoff for inclusion above an EDSS score of 5 or 5.5, with an average score across the trials ranging from 1.5 to 4. The majority of trials (71%) included treatment-naïve patients. Most studies (76%) included exclusively RRMS patients, six studies included ≥ 85% RRMS patients and five did not clearly report MS subtypes [31, 38,39,40,41]. Over one-fifth (22%) of trials were rated as low risk of bias in all domains, 46% had at least one domain at a high risk of bias (see ESM Appendix 4b–f). There was some variation in the follow-up time between studies. Many studies did not report one or more of the baseline details. The definition of relapse was variable; most studies defined it as new or worsening clinical signs or symptoms. However, the definition may also include neurological symptoms, with symptoms lasting for 24 h or up to 30 days and possibly but not definitively accompanied with fever and infection.
After comparison of the baseline data, 18 studies were excluded from the networks as they were judged to be unable to uphold similarity assumptions. The excluded studies are listed in ESM Appendix 4g with accompanying reasons for exclusion, which were: dose, follow-up time, outcome definition and MS type. For the remaining 33 studies—18 placebo-controlled and 15 head-to-head— four networks were created, one for each outcome of interest. The network for ARR (28 studies) is shown in Fig. 2 and the networks for CDP3M (13 studies), CDP6M (14 studies) and SAEs (6 studies) are shown in ESM Appendix 5. Looking at the networks, most comparisons involved one trial, some had two trials, but only two treatment comparisons had three trials. In the SAE network, data from CARE MS-I and CARE MS-II (alemtuzumab 12 mg once daily vs. IFN beta-1a 44 mcg three times per week) could not be connected to the network due to the absence of data to inform the intermediate connections.
Network diagram for annualised relapse rate (28 studies, with name of study given in uppercase letters on connecting lines). bid Twice daily, DMF dimethyl fumarate, GA glatiramer acetate, IFN interferon, PEG pegylated, q2w every 2 weeks, q24w every 24 weeks, q4w every 4 weeks, qad every other day, qd once daily, qw once per week, tiw three times per week
Analysis and Interpretation
Relapse rate was significantly reduced for all treatments compared with placebo. Treatments were ranked according to the probability of being the most effective (Fig. 3; Table 1; ESM Appendix 6). Alemtuzumab had the highest probability (63%) of being the most effective treatment, in terms of ARR compared with placebo (RR 0.28, 95% credible intervals [CrI] 0.21–0.38), followed by natalizumab (30% probability; RR 0.32, 95% CrI 0.23–0.43). All other treatments had < 10% probability of being most effective versus placebo, with IFN beta-1a, IFN beta-1b, GA 20 mg once daily and teriflunomide all having 0% probability. Pairwise comparisons were performed where possible to identify any potential heterogeneity (ESM Appendix 7a–e). High heterogeneity was identified in the comparison of GA 20 mg versus placebo (I2 = 86%). The 1987 Bornstein et al. study [31] appeared to be an outlier and was excluded in the sensitivity analyses for both pairwise meta-analysis and MTC analysis; exclusion of this study reduced heterogeneity (0%) and altered the RRs slightly while narrowing the 95% CrI (ESM Appendix 7f, g). Sensitivity analyses were also carried out to assess the impact of using a fixed-effects model and the impact of pooling follow-up data by analysing 24-month data only (ESM Appendix 7h, i); for both analyses, there were no major differences in the results. There was no evidence of statistically significant inconsistency using the node splitting method, or by comparison of the direct analyses with the MTC results (Fig. 3; ESM Appendix 7a–e, 7j).
There was substantial uncertainty associated with disease progression, likely due to the small number of studies informing each treatment comparison in the network. There was a reduction in the risk of both CDP3M and CDP6M for all treatments relative to placebo; however, the differences were statistically significant for only two and three treatments compared with placebo, respectively (CDP3M: alemtuzumab and ocrelizumab; CDP6M: alemtuzumab, ocrelizumab and natalizumab). Again, treatments were ranked according to the probability of being the most effective (Fig. 4 [CDP3M], Fig. 5 [CDP6M]; ESM Appendix 6). The results were numerically similar for the CDP3M and CDP6M analyses. In the case of disability progression at 6 months, only alemtuzumab, ocrelizumab and natalizumab were significantly more effective than placebo (Fig. 5). Alemtuzumab had the highest probability (45%) of being the most effective treatment compared with placebo (HR 0.37, 95% CrI 0.17–0.73), followed by ocrelizumab (26% probability; HR 0.40, 95% CrI 0.19–0.85) and then natalizumab (17% probability; HR 0.46, 95% CrI 0.25–0.84). There was no evidence of heterogeneity in any pairwise comparisons (ESM Appendices 8a, b and 9a, b). Sensitivity analyses were carried out to assess the impact of using a fixed-effects model in both CDP3M and CDP6M (ESM Appendices 8c and 9c), and the results were similar in both models. Analysis of inconsistency using the node splitting method was not possible due to a lack of studies.
There were no statistically significant differences in the odds of a SAE for any treatment in the network relative to placebo (Fig. 6; ESM Appendix 10). There was moderate evidence of heterogeneity (ESM Appendix 10a). Sensitivity analysis was carried out to assess the impact of using a fixed-effects model (ESM Appendix 10b) and found that the treatment effects were similar in both models. As the definition of SAE considered for inclusion in the network excluded MS relapse, we performed a sensitivity analysis to investigate the impact of its inclusion (ESM Appendix 10c). The results of this analysis showed that inclusion of MS relapse had little impact on the results for IFN beta-1a or fingolimod but it did reduce the estimated treatment effects relative to placebo for GA, DMF, and natalizumab. Analysis of inconsistency was not possible.
Discussion
Substantial differences in the conduct of previous systematic reviews in terms of the patient population included treatments and analysis methods, which mean that the results of those studies are not directly comparable with the results of this review.
This review performed Bayesian random-effects modelling to compare all treatments to each other and placebo, and the results were ranked according to the probability that each treatment was the most effective in terms of risk of relapse and disability progression. Sensitivity analyses were performed to investigate heterogeneity, using fixed-effects instead of random-effects models, pooling of follow-up data and the definition of SAE. All of these analyses indicated that the base–case analysis was robust, and we found no evidence of statistically significant inconsistency.
The networks presented here were clearly defined for the treatments at individual nodes, and different treatment regimens were not combined. We clearly presented patient and study characteristics. The process for including studies into the network and the reasons for exclusion were also clearly reported. The identified evidence base had variable diagnostic criteria, which reflected the clinical changes that have occurred over the 30-year time span covered by the review. Sormani et al. [17] discussed the importance of the similarity of the population in terms of diagnosis and MS subtypes. To ensure similarity we restricted patients to those with RRMS (≥ 85% RRMS). In addition, we restricted studies to those reporting SAEs (not adverse events [AEs] and not SAEs whose definition included MS relapse) to ensure that SAEs were consistent between studies. For similar reasons, CDP3M and CDP6M were analysed at 24-month follow-up while we combined all follow-up times greater than 12 months for ARR. A combination of follow-up times allowed a network to be created that compared more relevant treatments. A panel of expert MS clinicians was consulted to interpret this variability and confirmed that the studies included were clinically comparable and outcomes were clinically relevant. The clinical similarity of study populations was often unclear due to absent or heterogeneous reporting of key patient characteristics.
As treatments for MS have evolved over time, there has been a methodological shift from simple placebo-controlled studies in naïve patients to complex head-to-head or active-controlled studies, with or without a placebo reference, in a mixed population of treatment-naïve and previously treated patients. This trend will have an impact on the MTC network and, ultimately, the relative effect. For example, a head-to-head trial in patients who have experienced disease activity on previous therapy might introduce bias in the relative effects seen in such an MTC and could overestimate effects versus placebo. The varying ARR and progression rates observed with IFN beta-1a over many years in different studies with different comparators highlight the difficulty of this exercise. The defined networks presented here were limited by the lack of head-to-head comparisons between some treatments, which remains an unmet need in MS, and the limited number of studies per comparison. Given these limitations, MTC is the only way to conduct such cross-study comparisons.
While MTCs allow for the comparison of products where no direct evidence exists, it is important to note that they should not be used in isolation as a tool to assist decision making. Patient characteristics and preference, heterogeneity of disease course and unmet need should all be considered when looking into individual treatment decisions. MTCs provide estimates of comparative efficacy given the available data included. The further application of real-world data and comparisons employing propensity matching and adjustment have the possibility to enhance these MTC findings and expand evidence across populations beyond the RCT. Although at a risk of introducing greater heterogeneity, the convergence of evidence from different sources may help to provide more precision to conclusions in comparing across the treatment arena.
Strengths and Limitations
We identified a number of major challenges during MTC analysis which were inherent in the available evidence base for MS trials, including: reporting of AEs; lack of baseline details; sparse networks which can limit opportunities for sensitivity or subgroup analyses and defining similarity assumptions.
MS networks are sparse in comparison to other disease types, such as schizophrenia [42]. Many links in the network were formed from single studies; therefore, the exclusion of a single study for any sensitivity analysis could lead to the breakdown of the entire network. The reliance of the networks on limited studies prevented the investigation of subgroups of interest (MS subtypes, non-responders, treatment naïve patients, previous treatments) or further sensitivity analyses (risk of bias, disease severity, study size, use of an active comparator or placebo). Several studies did not report relevant data, such as standard deviation or CIs to allow inclusion in the MTC. It is a challenge within the design of a network (especially sensitivity analyses) to avoid the temptation to create the network at the expense of the quality of data by compromising similarity assumptions or data imputation.
There were no statistically significant differences in the odds of an SAE for any treatment in the network relative to placebo (Fig. 6; ESM Appendix 10), which may reflect limited statistical power due single studies informing the different arms of the network; alternatively, it may indicate that there are no differences between the treatments.
This systematic review followed best practice standards [20], and the methodology was clearly reported. There were no date or language restrictions. The searches included a comprehensive set of sources and databases that were designed to identify articles about the interventions or the comparators. Two independent reviewers were involved in all steps of the review, thereby ensuring low risk of meta bias [43]. It is important for decision making to include both components of the benefit–risk equation, and therefore we selected both efficacy and safety outcomes. However, alternative study designs are more appropriate for identifying AEs, and it is likely that restricting inclusion to RCTs compromised the evidence for SAEs in this review. This study may be improved by including non-randomised trials [21, 44, 45].
The best practice requirement to submit our MTC protocol to PROSPERO post-dated the data extraction phase of our initial analysis, thereby preventing us from registering our study. However, we recognise the importance of registration to PROSPERO in demonstrating transparency and therefore encourage all researchers to submit their MTC protocol to ensure that high standards of review work are upheld.
As discussed above, the clinical similarity of the included studies was difficult to assess due to unclear reporting of key information. We consulted a panel of clinicians to provide expert guidance on the similarity of the included studies. In cases where the available information was ambiguous, we favoured inclusion of the studies wherever possible in order to provide the best available evidence. Since many links in the network were formed from single studies, the alternative would be that networks could not be formed for several outcomes and the comparisons of interest could not be assessed. However, it should be considered that the factors outlined above may have introduced heterogeneity into the analysis.
The use of date limits can introduce bias into analysis and may compound the problems associated with sparse networks by excluding important evidence. Consistent with the Cochrane review standards [46], we did not apply a date limit and performed searches in each database from inception, with the aim of identifying all relevant evidence. However, we recognise that the inclusion of earlier studies may present a limitation in our analysis in that temporal changes in diagnostic criteria, treatment patterns and clinical trial populations could introduce further heterogeneity. In fact, when high heterogeneity was observed in ARR for comparisons of GA versus placebo, the exclusion of a 1987 study by Bornstein et al. [31] from the sensitivity analysis resulted in only a slight change in the point estimate compared to the base case.
Conclusions
Meta-analyses confirmed that all DMTs reduced ARR compared with placebo as defined by the included studies. This result generally aligns with other those from MTCs, as reported in 2020 by Sormani et al. [17], in which the estimated treatment effects relative to placebo were in the same direction for those treatment effects that were reported by multiple studies. This trend was consistent across outcomes and across populations, but the magnitude of the effects and the associated uncertainty varied as a result of the differences between studies [17]. In terms of ARR, alemtuzumab and natalizumab had the highest probabilities of being the most effective treatment compared with placebo.
Uncertainty in results for CDP3M and CDP6M were likely a result of the small number of studies that could be included in the analysis. However, the findings suggested that only the monoclonal antibodies alemtuzumab, ocrelizumab and natalizumab reduced the rate of disability progression at 6 months compared with placebo. Although data on SAEs were not available for all DMTs, the result of analysis implies that patients receiving active treatment with DMTs for RRMS experience fewer SAEs than those receiving placebo.
The rigorous and transparent approach taken to performing and reporting the current study allows the results to be held as a benchmark for comparisons with future new agents.
References
Caldwell DM. An overview of conducting systematic reviews with network meta-analysis. Syst Rev. 2014;3:109.
Tonin FS, Rotta I, Mendes AM, Pontarolo R. Network meta-analysis: a technique to gather evidence from direct and indirect comparisons. Pharm Pract. 2017;15(1):943.
Dias S, Welton NJ, Sutton AJ, Caldwell DM, Lu G, Ades AE. NICE DSU technical support document 4: inconsistency in networks of evidence based on randomised controlled trials. London: National Institute for Health and Care Excellence (NICE); 2014. http://nicedsu.org.uk/wp-content/uploads/2016/03/TSD4-Inconsistency.final_.15April2014.pdf. Accessed 03 Feb 2020.
Hutton B, Salanti G, Caldwell DM, et al. The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: checklist and explanations. Ann Intern Med. 2015;162(11):777–84.
Ades A, Caldwell DM, Reken S, Welton NJ, Sutton AJ, Dias S. NICE DSU technical Support document 7: evidence synthesis of treatment efficacy in decision making: a reviewers checklist. London: National Institute for Health and Care Excellence (NICE); 2012. http://nicedsu.org.uk/wp-content/uploads/2016/03/TSD7-reviewer-checklist.final_.08.05.12.pdf. Accessed 03 Feb 2020.
Hoaglin DC, Hawkins N, Jansen JP, et al. Conducting indirect-treatment-comparison and network-meta-analysis studies: report of the ISPOR Task Force on Indirect Treatment Comparisons Good Research Practices: part 2. Value Health. 2011;14(4):429–37.
Jansen JP, Fleurence R, Devine B, et al. Interpreting indirect treatment comparisons and network meta-analysis for health-care decision making: report of the ISPOR Task Force on Indirect Treatment Comparisons Good Research Practices: part 1. Value Health. 2011;14(4):417–28.
Richards RG, Sampson FC, Beard SM, Tappenden P. A review of the natural history and epidemiology of multiple sclerosis: implications for resource allocation and health economic models. Health Technol Assess. 2002;6(10):1–73.
Multiple Sclerosis International Federation (MSIF). Atlas of MS 2013. London: MSIF; 2013. https://www.msif.org/about-us/who-we-are-and-what-we-do/advocacy/atlas/. Accessed 03 Feb 2020.
Rae-Grant A, Day G, Marrie R, et al. Practice guideline recommendations summary: disease-modifying therapies for adults with multiple sclerosis: Report of the Guideline Development, Dissemination, and Implementation Subcommittee of the American Academy of Neurology. Neurology. 2018;90(17):777–88.
Poser CM, Paty DW, Scheinberg L, et al. New diagnostic criteria for multiple sclerosis: guidelines for research protocols. Ann Neurol. 1983;13(3):227–31.
McDonald WI, Compston A, Edan G, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50(1):121–7.
Polman CH, Reingold SC, Edan G, et al. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann Neurol. 2005;58(6):840–6.
Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292–302.
Zhang Y, Salter A, Wallström E, Cutter G, Stüve O. Evolution of clinical trials in multiple sclerosis. Therapeutic Adv Neurol Disorders. 2019;12(1). https://doi.org/10.1177/1756286419826547.
Song F, Loke YK, Walsh T, Glenny A-M, Eastwood AJ, Altman DG. Methodological problems in the use of indirect comparisons for evaluating healthcare interventions: survey of published systematic reviews. BMJ. 2009;338:b1147.
Sormani MP, Wolff R, Lang S, et al. Overview of differences and similarities of published mixed treatment comparisons on pharmaceutical interventions for multiple sclerosis. Neurol Ther. 2020. https://doi.org/10.1007/s40120-020-00213-4.
McCool R, Wilson K, Arber M, et al. Systematic review and network meta-analysis comparing ocrelizumab with other treatments for relapsing multiple sclerosis. Multiple Sclerosis Relat Disord. 2019;29:55–61.
National Institute for Health and Care Excellence (NICE). Single technology appraisal: User guide for company evidence submission template. London: NICE; 2017. https://www.nice.org.uk/process/pmg24/chapter/instructions-for-companies. Accessed 03 Feb 2020.
Higgins J, Thomas J, Chandler J, et al. Cochrane handbook for systematic reviews of interventions. Version 5.1.0. (updated March 2011). Cochrane. http://www.training.cochrane.org/handbook. Accessed 07 Aug 2014.
Centre for Reviews and Dissemination (CRD). Systematic reviews. CRDs guidance for undertaking reviews in health care. York: CRD; 2009. https://www.york.ac.uk/media/crd/Systematic_Reviews.pdf. Accessed 03 Feb 2020.
Jansen JP, Trikalinos T, Cappelleri JC, et al. Indirect treatment comparison/network meta-analysis study questionnaire to assess relevance and credibility to inform health care decision making: an ISPOR-AMCP-NPC Good Practice Task Force report. Value Health. 2014;17(2):157–73.
Higgins J, Altman DG, Gotzsche PC, et al. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. Br Med J. 2011;343.d5928. https://doi.org/10.1136/bmj.d5928.
Schwarzer G. Meta: general package for meta-analysis. R package version 4.3-0. 2015. https://cran.r-project.org/web/packages/meta/meta.pdf. Accessed 28 Jan 2016.
R Core Team. R: a language and environment for statistical computing. Copenhagen: Global Biodiversity Information Facility; 2015. https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing. Accessed 28 Jan 2016.
van Valkenhoef G, Kuiper J. gemtc: network meta-analysis using Bayesian methods. R package version 0.7-1. 2015. https://cran.r-project.org/web/packages/gemtc/gemtc.pdf. Accessed 13 Oct 2017.
Brooks S, Gelman A. General methods for monitoring convergence of iterative simulations. J Comput Graph Stat. 2012;7(4):434–55.
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16.
Suthar A, Lawn S, del Amo J, et al. Antiretroviral therapy for prevention of tuberculosis in adults with HIV: a systematic review and meta-analysis. PLoS Med. 2012;9(7):e1001270.
Woods BS, Hawkins N, Scott DA. Network meta-analysis on the log-hazard scale, combining count and hazard ratio statistics accounting for multi-arm trials: a tutorial. BMC Med Resour Methodol. 2010;10(1):54.
Bornstein M, Miller A, Slagle S, et al. A pilot trial of Cop 1 in exacerbating-remitting multiple sclerosis. N Engl J Med. 1987;317(7):408–14.
Gobbi C, Meier DS, Cotton F, et al. Interferon beta 1b following natalizumab discontinuation: one year, randomized, prospective, pilot trial. BMC Neurol. 2013;13(1):101.
Mokhber N, Azarpazhooh A, Orouji E, et al. Cognitive dysfunction in patients with multiple sclerosis treated with different types of interferon beta: a randomized clinical trial. J Neurol Sci. 2014;342(1–2):16–20.
Mokhber N, Azarpazhooh A, Orouji E, et al. Therapeutic effect of Avonex, Rebif and Betaferon on quality of life in multiple sclerosis. Psychiatry Clin Neurosci. 2015;69(10):649–57.
Patti F, L’Episcopo M, Cataldi M, Reggio A. Natural interferon-β treatment of relapsing—remitting and secondary-progressive multiple sclerosis patients. A two-year study. Acta Neurol Scand. 1999;100(5):283–9.
Romine JS, Sipe JC, Koziol JA, Zyroff J, Beutler E. A double-blind, placebo-controlled, randomized trial of cladribine in relapsing-remitting multiple sclerosis. Proc Assoc Am Physicians. 1999;111(1):35–44.
Stelmasiak Z, Solski J, Nowicki J, Jakubowska B, Ryba M, Grieb P. Effect of parenteral cladribine on relapse rates in patients with relapsing forms of multiple sclerosis: results of a 2-year, double-blind, placebo-controlled, crossover study. Multiple Sclerosis. 2009;15(6):767–70.
O’Connor P, Filippi M, Arnason B, et al. 250 microg or 500 microg interferon beta-1b versus 20 mg glatiramer acetate in relapsing-remitting multiple sclerosis: a prospective, randomised, multicentre study. Lancet Neurol. 2009;8(10):889–97.
Crentsil C, Scolding N, Wilkins A, Burrow J, Bennetto L, Ingles K. A comparison of the efficacy of interferon-beta and glatiramer acetate in relapse-rate reduction: a prospective randomisation study. Paper presented at 28th Congress of the European Committee for Treatment and Research in Multiple Sclerosis; 10–12 Oct 2012; Lyon, France. Multiple Sclerosis. 2012;18(4 Suppl 1):209.
Kira J, Itoyama Y, Kikuchi S, Hao Q, Kurosawa T, Nagato K. Oral fingolimod (FTY720) in Japanese patients with relapsing multiple sclerosis: results of a 12-month, phase 2 extension study. Paper presented at 5th Joint Triennial Congress of the European and Americas Committees for Treatment and Research in Multiple Sclerosis; 19–22 Oct 2011; Amsterdam: the Netherlands. Multiple Sclerosis. 2011;17(10 Suppl 1):S193.
PRISMS (Prevention of Relapses and Disability by Interferon beta-1a Subcutaneously in Multiple Sclerosis) Study Group. Randomised double-blind placebo-controlled study of interferon beta-1a in relapsing/remitting multiple sclerosis. Lancet. 1998;352(9139):1498–504.
Leucht S, Cipriani A, Spineli L, et al. Comparative efficacy and tolerability of 15 antipsychotic drugs in schizophrenia: a multiple-treatments meta-analysis. Lancet. 2013;382(9896):951–62.
Whiting P, Savović J, Higgins JPT, et al. ROBIS: a new tool to assess risk of bias in systematic reviews was developed. J Clin Epidemiol. 2016;69:225–34.
Golder S, Loke YK, Bland M. Meta-analyses of adverse effects data derived from randomised controlled trials as compared to observational studies: methodological overview. PLoS Med. 2011;8(5):e1001026.
Loke YK, Golder SP, Vandenbroucke JP. Comprehensive evaluations of the adverse effects of drugs: importance of appropriate study selection and data sources. Therap Adv Drug Saf. 2011;2(2):59–68.
Higgins J, Lasserson T, Chandler J, et al. Methodological expectations of Cochrane Intervention Reviews. Searching for studies—Standard C35. Cochrane Community. https://community.cochrane.org/mecir-manual/standards-conduct-new-cochrane-intervention-reviews-c1-c75/performing-review-c24-c75/searching-studies-c24-c38. Accessed 03 Feb 2020.
Acknowledgements
Richard Birnie produced the tables and figures, screened the search results, selected full-text studies for inclusion and performed data extraction.
Funding
Work on this systematic review was sponsored by Biogen, and Biogen was given the opportunity to comment on the draft paper. All authors gave final approval of the version to be published. Biogen funded the journal’s Rapid Service Fee.
Medical Writing and Editorial Assistance
Editorial assistance in the preparation of this article was provided by Zoe Whittington and James Davies of OPEN VIE (Marlow, UK). Funding for this assistance was provided by Biogen.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Gavin Giovannoni, Shona Lang, Robert Wolff, Steven Duffy, Robert Hyde, Elizabeth Kinter, Craig Wakeford, Maria Pia Sormani and Jos Kleijnen have nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Data Availability
All data generated or analyzed during this study are included in this published article/as supplementary information files.
Author information
Authors and Affiliations
Corresponding author
Additional information
Digital Features
To view digital features for this article go to https://doi.org/10.6084/m9.figshare.12912446.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Giovannoni, G., Lang, S., Wolff, R. et al. A Systematic Review and Mixed Treatment Comparison of Pharmaceutical Interventions for Multiple Sclerosis. Neurol Ther 9, 359–374 (2020). https://doi.org/10.1007/s40120-020-00212-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40120-020-00212-5