Abstract
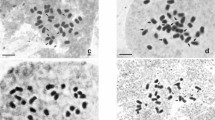
Karyomorphological analysis was done in cooking banana cultivars (Musa acuminate L.) namely cv. Gajabnatala, cv. Paunsiabnatala, cv. Mendhibantala, cv. Desi-Dakshinisagar, cv. Normal-Daksinisagar, cv. Mutant-Daksinisagar and cv. Shankara of Odisha, India. All cultivars were 2n = 3x = 33 with an average mitotic metaphase chromosome length ranged from 1.20 µm in cv. Shankara to 2.15 µm in cv. Desi-Dakhinisagara. Total chromosome length varied from 39.75 to 71.04 µm. Predominance of nearly median chromosomes found a typical characteristic of the seven studied plantain cultivars in which the total F% varied from 38.63 in cv. Mendhibantala to 45.31 in cv. Desi-Dakhinisagara. The total chromosome volume was found lowest in cv. Shankara (18.46 µm) and highest in cv. Desi-Dakhinisagara (32.24 µm). The interphase nuclear volume calculated ranged from 562.50 µm3 in cv. Shankara to 1463.04 µm3 in cv. Mendhibantala. ‘Desi-Dakhinisagara’ with the highest karyotype asymmetry value found advanced cultivar among all with lowest asymmetry in cv. Mutant Dakhinisagara found primitive. UPGMA clustering of five intra-chromosomal and two inter-chromosomal asymmetry indexes revealed the plantain cultivars except cv. Mendhibantala along with cv. Shankara and cv. Dakhinisagara along with cv. Mendhibantala were grouped together in the phylogenetic tree.




Similar content being viewed by others
References
Robinson JC (1996) Bananas and Plantains. CAB International, Wallingford
FAO (2003) The world banana economy 1985–2002. Economic and Social Department, Food and Agriculture Organization, Rome. FAOSTAT. Various years. www.faostat.fao.org.
Cheesman EE (1947) Classification of the bananas. I. The genus Ensete horan and the genus Musa L. Kew Bull 2:97–117
Argent G (1976) The wild bananas of papua new guinea. Notes Roy Bot Gard Edinb 35:77–114
Simmonds NW, Shepherd K (1955) The taxonomy and origins of the cultivated bananas. Bot J Linn Soc 55:302–312
Dolezel J, Dolezelova M, Novak FJ (1994) Flow cytometric estimation of nuclear DNA amounts in diploid bananas (Musa acuminata and M. balbisiana). Biol Plant 36:351–357
D’Hont A, Paget-Goy A, Escoute J, Carreel F (2000) The interspecific genome structure of cultivated banana, Musa spp. revealed by genomic DNA in situ hybridization. Theor Appl Genet 100:177–183
Osuji JO, Okuli BE, Hilary OE (2006) Karyotypes of the A and B genomes of Musa L. Cytologia 71:21–24
Manzo-Sanchez G, Buenrostro-Nava MT, Guzman-Gonzalez S, Orozco-Santos M, Youssef M, Medrano RMEG (2015) Genetic diversity in bananas and plantains (Musa spp.). In: Caliskan M, Oz GC, Kavakli IH, Ozcan B (Eds.), Molecular Approaches to Genetic Diversity, Tech, Rijeka, Croatia, pp 93–121
Stebbins GL (1971) Chromosomal evolution in higher plants. Edward Arnold (Publishers) Ltd, London, UK
Peruzzi L, Eroğlu HE (2013) Karyotype asymmetry: again, how to measure and what to measure? Comp Cytogenet 7:1–9
Das AB, Mallick R (1993) Karyotype diversity and interspecific 4C DNA variation in Bupleurum. Biol Plant 35:355–363
Das AB, Das P (1997) Estimation of nuclear DNA and karyotype analysis in nine cultivars of Musa acuminata. Cytobios 90:181–192
Paszko B (2006) A critical review and a proposal of karyotype asymmetry indices. Plant Syst Evol 258:39–48
Eroğlu HE (2015) Which chromosomes are subtelocentric or acrocentric? A new karyotype symmetry/asymmetry index. Caryologia 68:239–245
Garcia-Vallve S, Palau J, Romeu A (1999) Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Bol Evol 16:1125–1134
OrtizR SR (2014) From cross breeding to biotechnology-facilitated improvement of banana and plantain. Biotechnol Adv 32:158–169
Mantell SH, Matthews JA, McKee RA (1985) Principles of plant biotechnology: an introduction to genetic engineering in plants. Blackwell Scientific Publications, London
Hartwell LH, Hood L, Goldberg ML, Reynolds AE, Silver LM, Veres RC (2000) Genetics: from gene to genomes. McGraw-Hill, Boston, pp 731–751
Leven A, Fredya K, Sandberg A (1964) Nomenclature for centromeric position on chromosome. Heredity 52:201–220
Seijo JG, Fernádez A (2003) Karyotype analysis and chromosome evolution in South American species of Lathyrus (Leguminosae). Am J Bot 90:980–987
Šimoníková D, Němečková A, Karafiátová M, Uwimana B, Swennen R, Doležel J, Hřibová E (2019) Chromosome painting facilitates anchoring reference genome sequence to chromosomes in situ and integrated karyotyping in banana (Musa Spp.). Front Plant Sci. https://doi.org/10.3389/fpls.2019.01503
Hřibová E, Imoníková D, Doležel J (2020) Molecular cytogenetics and diversity in Musa. In: Abstract volume of International Conference on Banana-2020 on ‘Innovations in sustainable production and value chain management in banana’, NRC Banana, Tiruchirappalli, Tamil Nadu, pp 20
Ghosh S, Das A, Ghorai A, Jha TB (2013) Comparative karyomorphology of edible Musa cultivars of West Bengal. Caryologia 66:243–250
Sharrock S (1998) Collecting the Musa gene pool in Papua New Guinea. In: Guarino L, Rao VR, Reid R (eds) Collecting plant genetic diversity. CAB International, Wallingford, pp 647–658
Stace CA (2000) Cytology and cytogenetics as a fundamental taxonomic resource for the 20th and 21st centuries. Taxon 49:451–477
Romero-Zarco CR (1986) A new method for estimating karyotype asymmetry. Taxon 35:526–530
Peruzzi L, Leitch IJ, Caparelli KF (2009) Chromosome diversity and evolution in Liliaceae. Ann Bot 103:459–475
Acknowledgements
The research was supported with funds by Department of Biotechnology, Govt. of India from the project (DBT-NER/AGRI/33/2016 (Group-I, Application No. 90). We are thankful to the Head of the Botany, Utkal University, for providing supportive and microscopic laboratory facilities, which are funded by DRS-SAP-III, University Grant Commission and DST-FIST, Govt. of India, to carry out the experiment. ABD acknowledges the receipt of Emeritus Professorship from CSIR, HRDG EMR-II (Scheme No. 21 (1107) /20/EMR-II), Ministry of Science and Technology, Govt. of India . All authors declare that they have checked the text of the manuscript, tables and photographs provided out of our research work and approve the submission of the manuscript to the Journal for publication.
Author information
Authors and Affiliations
Contributions
Prof. ABD conceives the experiments and finalizes the MS text write-up as principal investigator of DBT, and CSIR EMR-II, DST, Govt. of India Project. Mrs. EP took the experimental data and part of calculation as JRF of the Project. Mr. SD and Dr. CP were responsible for data calculation, graph preparation and first draft manuscript writing as JRF and co-principal investigator of the DBT Project respectfully.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance Statement
Genetic improvement of cooking banana is of greater importance as it is one of the major carbohydrate sources of millions of poor people with iron, magnesium, calcium minerals and for dietary fibers. Chromosome morphology, karyotype asymmetry of seven cooking cultivars of Musa acuminate showed triploid (2n = 3x = 33) chromosome. Variation in intra-chromosomal karyotype asymmetry established phylogenetic relationships among cooking banana which may be used for utilization of disease resistant characters to table-top banana in crop improvement through breeding.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Panda, E., Dehery, S.K., Pradhan, C. et al. Study of Karyotype Asymmetry and Chromosome Number in Seven Cooking Bananas (Musa acuminata L.). Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 93, 387–395 (2023). https://doi.org/10.1007/s40011-022-01427-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-022-01427-2