Abstract
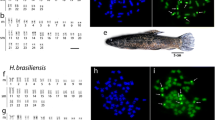
Snakehead fishes are important food fishes from freshwater ecosystems with their significant contribution in capture fisheries. They also have high potential for aquaculture. Cytogenetic variability between the Channa punctata and C. striata was assessed in the present investigation. The study revealed diploid chromosome number, karyotype configuration and fundamental arm numbers as 32, 18m + 10sm + 4st and 64, respectively, in C. punctata and 40, 6m + 2sm + 10st + 22t and 68 in C. striata without any heteromorphic pair(s) in both the species. Rhodamine-labeled 18S rDNA probe was constructed and hybridized on the metaphase chromosome spreads of both the species. The signals of the hybridized probe were observed on two sub-metacentric and two sub-telocentric chromosome pairs in C. punctata and C. striata, respectively. The study revealed that 18S ribosomal genes are present on morphologically different chromosome pairs which have evolved by means of chromosome fragmentation and their rearrangements during the evolution of the species. The information is useful for 18S gene-bearing chromosomes identification, in cytotaxonomy and documentation of the hybrids of the two species.


Similar content being viewed by others
References
Fricke R, Eschmeyer WN, van der RL (eds) (2020) Eschmeyer's catalog of fishes: genera, species, references. http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp. Electronic version Accessed 05.02.2020
Jhingran VG (1982) Fish and Fisheries of India. 70. Publ. Hindus. Publish. Corpo. Delhi, India, pp 467–470
Michelle NYT, Shanti G, Loqman MY (2004) Effect of orally administered Channa striatus extract against experimentally-induced osteoarthritis in rabbits. Int J Appl Res Vet Med 2:171–175
Hossain MK, Latifa GA, Rahman MM (2008) Observations on induced breeding of snakehead murrel, Channa striatus (Bloch, 1793). Int J Sustain Crop Prod 3:65–68
Kumar R, Baisvar VS, Kushwaha B, Waikhom G, Singh M (2019) Evolutionary analysis of genus Channa based on karyological and 16S rRNA sequence data. J Genet 98:112
Baisvar VS, Kumara R, Singh M, Singh AK, Chauhan UK, Mishra AK, Kushwaha B (2018) Genetic diversity analyses for population structuring in Channa striata using mitochondrial and microsatellite DNA regions with implication to their conservation in Indian waters. Meta Gene 16:28–38
Bhat AA, Haniffa MA, Divya PR, Gopalakrishnan A, Milton MJ, Kumar R, Paray BA (2012) Molecular characterization of eight Indian Snakehead species (Pisces: Perciformes Channidae) using RAPD markers. Mol Biol Rep 39:4267–4273
Diniz D, Laudicina A, Bertollo LAC (2009) Chromosomal location of 18S and 5S rDNA sites in Triportheus fish species (Characiformes, Characidae). Genet Mol Biol 32:37–41
Meyer A, Todt C, Mikkelsen NT, Lieb B (2010) Fast evolving 18S rRNA sequences from Solenogastres (Mollusca) resist standard PCR amplification and give new insights into mollusk substitution rate heterogeneity. BMC Evol Biol 10:70–81
Chenuil A (2006) Choosing the right molecular genetic markers for studying biodiversity: from molecular evolution to practical aspects. Genetica 127:101–120
Talwar PK, Jhingran AG (1991) Inland fishes of India and adjacent countries I. Oxford and IBH Publishing Co., NewDelhi
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Darriba D, Taboada GL, Doallo R, Posada D (2012) J Model Test 2: more models, new heuristics and parallel computing. Nat Methods 9:772
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Hadziavdic K, Lekang K, Lanzen A, Jonassen I, Thompson EM, Troedsson C (2014) Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS ONE 9:e87624. https://doi.org/10.1371/journal.pone.0087624
Kumar R, Kushwaha B, Nagpure NS, Behera BK, Srivastava SK, Lakra WS (2009) Physical mapping of rRNA gene in endangered fish Osteobrama belangeri (Valenciennes, 1844) (Family: Cyprinidae). Indian J Exp Biol 47:597–601
Gornung E (2013) Twenty years of physical mapping of major ribosomal RNA genes across the teleosts: a review of research. Cytogenet Genome Res 141:90–102
Kumar R, Baisvar VS, Kushwaha B, Waikhom G, Thoidingjam L, Singh SS (2020) Cytogenetic studies in Glossogobius giuris (Hamilton, 1822) through NOR-staining and FISH. Proc Natl Acad Sci India Sect B Biol Sci 90:221–226
Singh M, Kumar R, Nagpure NS, Kushwaha B, Mani I, Chauhan UK, Lakra WS (2009) Population distribution of 45S and 5S rDNA in golden mahseer, Tor putitora: population-specific FISH marker. J Genet 88:315–320
Singh M, Kumar R, Nagpure NS, Kushwaha B, Mani I, Lakra WS (2009) Extensive NOR site polymorphism in geographically isolated populations of golden mahaseer, Tor putitora. Genome 52:783–789
Acknowledgements
The authors are highly thankful to the DBT, New Delhi, for financial support to conduct the experiment and to the Director, ICAR-National Bureau of Fish Genetic Resources, Lucknow, for providing the laboratory facilities and other supports.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest related to this article. Further, both Basdeo Kushwaha and Vishwamitra Singh Baisvar may be considered as the first author in the manuscript due to their equal contributions.
Ethical Approval
For the present work, healthy live specimens of C. punctata and C. striata were collected from Gomti river near Lucknow, Uttar Pradesh, INDIA. All applicable national and institutional guidelines for the care and use of fishes were followed. The undertaken species is not listed in the endangered/ special category or otherwise enlisted fish species and does not restrict the use of this species for laboratory experiments/ research. The experiments with live fish do not come under the CPCSEA (Committee for the Purpose of Control and Supervision on Experiments on Animals) guidelines of the Ministry of Environment, Forest & Climate Change (MoEFCC), Government of India, New Delhi. There are no guidelines available for using live fish for experiments in India as on today.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance Statement: Channa punctata and C. striata are important food fishes and potential aquaculture species. The molecular cytogenetic information revealing the position of 18S rDNA through FISH will be useful in their cytotaxonomy, conservation and management.
Rights and permissions
About this article
Cite this article
Kushwaha, B., Baisvar, V.S. & Kumar, R. 18S rDNA mapping revealed conservation and rearrangements of chromosome segments in two Channa species. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 91, 675–679 (2021). https://doi.org/10.1007/s40011-021-01257-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-021-01257-8