Abstract
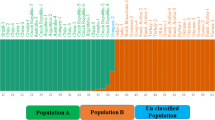
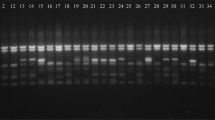
Common bean (Phaseolus vulgaris L.) is a vital legume used as human food, source of cash income, feed for livestock, and it increases the fertility of soil by its ability to fix nitrogen. The aim of this research was to determine the genetic diversity and population structure of 40 common bean germplasm cultivated in Kenya, using peroxidase gene (POX)-based molecular markers. The loci analyzed showed high diversity and amplified 624 alleles, ranging from 3 to 9 on every locus, with an average of 7.20. The PIC of the POX markers varied from 0.6204 to 0.9110, with an average of 0.7677. The range of the observed heterozygosity was from 0.6667 to 0.9150 with a mean of 0.7945, while the values of the mean genetic diversity ranged from 0.3072 to 0.4425 with a mean of 0.3972. The UPGMA phenogram separated the genotypes into two main genetic clusters, and the genotypes showed no grouping by geographical origins. The highest value of genetic variation was observed between the genotypes obtained from Western, Rift valley and Central regions of Kenya. Population structure analysis grouped the germplasm into 7 gene pools and showed that the genotypes have a common genetic lineage. AMOVA revealed higher genetic diversity (99%) within population than among population (1%), and this offers a reliable base for the design of genetic improvement schemes. Results of the present study can be used in future breeding programs and for the genetic improvement in common bean.





Similar content being viewed by others
References
Rezende AA, Pacheco MTB, Silva VSN, Ferreira TAPC (2018) Nutritional and protein quality of dry Brazilian beans (Phaseolus vulgaris L.). Food Sci Technol 38:421–427
Blair MW, Paulo I, Astudillo C, Grusak MA (2013) A legume biofortification quandary: variability and genetic control of seed coat micronutrient accumulation in common beans. Front Plant Sci 10:3389
Sajad MZ, Muslima N, Vandna R, Martin H, Ganesh KA, Randeep R (2015) Towards a common bean proteome atlas: looking at the current state of research and the need for a comprehensive proteome. Front Plant Sci 6:201
Valentini G, Gonçalves-Vidigal MC, Elias JCF, Moiana LD, Mindo NNA (2019) Population structure and genetic diversity of common bean accessions from Brazil. Plant Mol Biol Rep 36:5–6
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H (2018) DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip 32(2):261–285
Svetleva D, Pereira G, Carlier J, Cabrita L, Leitao J, Genchev D (2006) Molecular characterization of Phaseolus vlugaris L. genotypes included in Bulgarian collection by ISSR and AFLP analyses. Sci Hort 109:198–206
Nodari RO, Koinange EM, Kelly JD, Gepts P (1992) Towards an integrated linkage map of common bean: development of genomic DNA probes and levels of restriction fragment length polymorphism. Theor Appl Genet 84:186–192
Marotti I, Bonetti A, Minelli M, Catizone P, Dinelli G (2007) Characterization of some Italian common bean (Phaseolus Vulgaris L.) landraces by RAPD, semi-random and ISSR molecular markers. Genet Resour Crop Evol 54(1):175–188
Asfaw A, Blair MW, Almekinders C (2009) Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from the East African highlands. Theor Appl Genet 120:1–12
Gyang PJ, Nyaboga EN, Muge EK (2017) Molecular characterization of common bean (Phaseolus vulgaris L.) genotypes using microsatellite markers. J Adv Biol Biotechnol 13(2):1–15
Nemli S, Kaya HB, Tanyolac B (2014) Genetic assessment of common bean (Phaseolus vulgaris L.) accessions by peroxides gene-based markers. J Sci Food Agric 94(8):1672–1680
Collard BCY, Mackill DJ (2009) Conserved DNA-derived polymorphism (CDDP): a simple and novel method for generating DNA markers in plants. Plant Mol Biol Rep 27:558–562
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen JPT, Hyvönen J (2013) Advances in plant gene-targeted and functional markers: a review. Plant Methods 9:6
Gulsen O, Kaymak S, Ozongun S, Uzun A (2010) Genetic analysis of Turkish apple germplasm using peroxidase gene-based markers. Sci Hortic 125:368–373
Pinar H, Unlu M, Ercisli S, Uzun A, Bircan M (2016) Genetic analysis of selected almond genotypes and cultivars grown in Turkey using peroxidase-gene-based markers. J For Res 27(4):747–754
Ocal N, Mikail A, Osman G, Halit Y, Ilknur S, Nebahat S (2014) Genetic diversity, population structure and linkage disequilibrium among watermelons based on peroxidase gene markers. Sci Hortic 176:151–161
Welinder KG, Justesen AF, Kjaersgard IVH, Jensen RB, Rasmussen SK, Jespersen HM, Duroux L (2002) Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur J Biochem 269:6063–6081
Gulsen O, Shearman RC, Heng-Moss TM, Mutlu N, Lee DJ, Sarath G (2007) Peroxidase gene polymorphism in buffalograss and other grasses. Crop Sci 47:767–772
Yeh FC, Yang R (1999) Popgene version 1.31 Microsoft window-based freeware or population genetic analysis
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32(3):314–331
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76(10):5269–5273
Prichard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genet 155:945–959
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Wittayawannakul W, Garcia RN, Yllano OB, Borromeo TH, Namuco LO, Tecson-Mendoza EM (2010) Assessment of genetic diversity in Garciniaspecies using peroxidase, RAPD and gene sequence specific amplification polymorphism (GSSAP). Philipp Agric Sci 93:31–41
Santalla M, Rodin˜o A, de Ron A (2002) Allozyme evidence supporting southwestern Europe as a secondary center of genetic diversity for the common bean. Theor Appl Genet 104:934–944
Okii D, Tukamuhabwa P, Kami J, Namayanja A, Paparu P, Ugen M, Gepts P (2014) The genetic diversity and population structure of common bean (Phaseolus vulgaris L.) germplasm in Uganda. Afr J Biotechnol 13:2935–2949
Fisseha Z, Tesfaye K, Dagne K, Blair MW, Harvey J, Kyallo M, Gepts P (2016) Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) germplasm of Ethiopia as revealed by microsatellite markers. Afr J Biotechnol 15(52):2824–2847
Buah S, Buruchara R, Okori P (2017) Molecular characterisation of common bean (Phaseolus vulgaris L.) accessions from Southwestern Uganda reveal high levels of genetic diversity. Genet Resour Crop Evol 64:1–14
Rebaa F, Abid G, Aouida M, Abdelkarim S, Aroua I, Muhovski Y, Baudoin J, Mahmoud M, Sassi K, Jebara M (2017) Genetic variability in Tunisian populations of faba bean (Vicia faba L. var. major) assessed by morphological and SSR markers. Physiol Mol Biol Plants 23(2):397–409
Badiane FA, Gowda BS, Cissé N, Diouf D, Sadio O, Timko MP (2012) Genetic relationship of cowpea (Vigna unguiculata) varieties from Senegal based on SSR markers. Genet Mol Res 11(1):292–304
Acknowledgements
The authors would like to thank the Department of Biochemistry, University of Nairobi (UoN), for supporting this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest to publish this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance Statement The findings of the present study demonstrate the existence of a considerable amount of genetic variability among common bean genotypes grown in Kenya. This indicates the potential application of these genotypes in future breeding programs for genetic improvement of common beans.
Rights and permissions
About this article
Cite this article
Gyang, P.J., Muge, E.K. & Nyaboga, E.N. Genetic Diversity and Population Structure of Kenyan Common Bean (Phaseolus vulgaris L.) Germplasm Using Peroxidase Gene Markers. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 90, 293–301 (2020). https://doi.org/10.1007/s40011-019-01101-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-019-01101-0