Abstract
Research endeavors focused on examining genetic diversity play a pivotal role in advancing agricultural practices, as they furnish valuable insights for breeding programs and the preservation of germplasm. Wheat holds significant global importance as a cereal crop, serving as a primary source of sustenance for almost one-third of the global human population. In the current study, a total of 80 bread wheat genotypes were characterized using the start codon targeted (SCoT) marker system. A set of 14 polymorphic primers yielded a cumulative count of 399 bands with an average of 28.05 bands per primer. The range of diversity markers, such as polymorphism information content (0.26–0.59), effective numbers of alleles (1.27–1.59), Shannon’s information index (0.30–0.51), and gene diversity (0.18–0.34), provided evidence of elevated genetic differences within the examined germplasm. According to the findings of the molecular variance analysis, greater genetic variations are found within populations (92%). The model-based structural approach partitioned the 80 bread wheat genotypes into two distinct populations (Population A and B). The diversity indices based on the structures of the populations revealed that population A had greater diversity compared to population B. Both the principal coordinate analysis and the neighbor-joining analysis also differentiated the 80 different bread wheat genotypes based on their respective collection points. The present crop diversity offers great opportunities for enhancing crop quality and increasing tolerance to environmental challenges, both of which will lead to a reduction in the problem of food insecurity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite the appreciable progress made in the last decades aimed at reducing hunger, more than 800 million individuals globally experience persistent undernourishment (WFP 2020) According to a report shared by FAO (2015), approximately two billion people are facing essential micronutrient deficiencies called “hidden hunger”. Based on the presented data, ensuring food security emerges as a critical concern that requires immediate attention. This issue poses a significant threat to the fulfillment of sustainable development goals and gives rise to a range of economic and non-economic challenges (Battersby et al. 2017). Food security occurs if all people, at all times, have economic and physical access to sufficient, safe, and nutritious food that fulfills their dietary requirements for health and active life (FAO 2008). Climate change, increasing population, urbanization, loss of biodiversity, and industrialization are key factors playing a significant role in global hunger. Climate change poses the main threat to food security by affecting agricultural production through unprecedented biotic and abiotic stresses. According to Zhao et al. (2017), approximately a 3.1%–7.4% decline in production has been recorded for various important crops for each degree-Celsius increase in global mean temperature due to climate change. Besides climate changes, the rapidly growing population has become a key concern in present world, and the demand for this rapidly increasing population is a bitter and challenging question for the present breeders.
To feed 9.7 billion people by 2050, the Food and Agriculture Organization of the United Nations (FAO) has stated that there is a need to increase agricultural production by 60% compared to the current level. The scientific community, especially plant breeders, bears a significant responsibility to develop climate-resilient yet high-yielding and highly nutritious crop cultivars. This is crucial in addressing climate change, global hunger, andhidden hunger issues simultaneously. To achieve these objectives, it is imperative to effectively utilize and evaluate plant genetic diversity through germplasm characterization (Baloch et al. 2022; Baloch and Nadeem 2022; Mafakheri et al. 2020; Tabasum et al. 2020; Nadeem 2021). Genetic diversity assessment can be conducted through morphological, biochemical, and molecular markers; however, molecular markers are preferred due to their precision, reliability, reproducibility, and resilience to environmental factors (Nadeem et al. 2018). Different types of molecular markers (SSR, simple sequence repeat; AFLP, amplified fragment length polymorphism; DArT, diversity arrays technology; ISSR, inter simple sequence repeats; and RAPD, randomly amplified polymorphic DNA) have been developed and successfully applied in the marker-assisted breeding program of various crops. These markers are utilized for genetic population analysis, mapping quantitative trait loci (QTL), association analysis of various yield characteristics, and the detection of candidate genes.
Collard and Mackill (2009) developed a new marker system called start codon targeted (SCoT) polymorphism. This marker system is based on the short-conserved region flanking the ATG start codon in plant genes. It is a polymerase chain reaction (PCR) based marker system through which an 18-mer single primer targets the short consensus conserved region flanking the ATG translation initiation codon in plant genes (Collard and Mackill 2009). PCR products resulting from PCR are resolved using standard agarose gel electrophoresis (Guo et al. 2012). This marker system has been found to be cost-effective, resulting in high polymorphism. It is similar to ISSR and RAPD, is easy to use and its primers are universal in plants (Guo et al. 2012; Xiong et al. 2011). This marker has been successfully used in genetic diversity and population structure investigations of various plant species (Zarei et al. 2021; Yilmaz and Ciftci 2021; Tabasi et al. 2020; Al-Qurainy et al. 2015; Guo et al. 2012; Xiong et al. 2011).
Bread wheat (Triticum aestivum L.) is one of the most important crops, providing 20% of dietary energy and protein consumption worldwide (Braun et al. 2010). The present hexaploid (bread wheat) resulted from a hybrid cross between Triticum. dicoccoides and Aegilops tauschii (Velu et al. 2019). In the year 2020, a total land area of 219,006,893 hectares was used for the cultivation of wheat on a global scale, which ultimately yielding a production of 760,925,831 tons (Salama et al. 2022). To feed 9.7 billion people by 2050, it is estimated that wheat production should increase by 1.7% per year (Leegood et al. 2010). This can be achieved through the development of climate-resilient and high-yielding wheat cultivars. According to the published report (2019) from the IGC (International Grains Council), the global need for wheat is estimated to increase by one billion tons over the next four years. It appears that conventional breeding programs may be sufficient to meet this growing demand (Leśniowska-Nowak et al. 2021). However, breeders express significant concern that ongoing breeding cycles are resulting in a limited genetic diversity within cultivated genotypes. This hindrance in the gene pool poses a noteworthy challenge for future breeding programs, as the remaining variability might not be sufficient (Hegde et al. 2002). Molecular breeding with SCoT markers is a reliable approach for assessing the genetic variability among wheat cultivars (Hamidi et al. 2014) and selecting superior cultivars for further breeding programs (Yalinkiliç et al. 2024). Recently, various investigations worldwide have reported the application of SCoT markers test the diversity in wheat germplasm (Mohamed et al. 2017; El-Moneim et al. 2020; Shaban et al. 2022; Abouseada et al. 2023). Abouseada et al. (2023) investigated the genetic diversity in wheat cultivars using ISSR and SCoT primers with chloroplast DNA barcoding and grain scanning electron microscope (SEM). The investigation highlighted the potential of molecular markers and SEM in characterizing wheat diversity for breeding purposes. The results provided valuable insights into wheat genetic resources, aiding future breeding strategies for improved wheat cultivars. Furthermore, Shaban et al. (2022) assessed the genetic diversity in Egyptian wheat genotypes under drought and watering conditions using ISSR, SCoT and SDS-PAGE techniques. Significant variations were observed during data analysis, with various genotypes showing higher tolerance to drought stress. The researchers concluded that the findings can contribute to wheat breeding programs, enhance resilience to drought and ensure food security in changing climates. In a separate study Alshehri (2019) identified the genetic variation of Saudi wheat genotypes through SCoT and ISSR markers. Their findings contributed to a better understanding of wheat diversity and aided in developing resilient and high-yielding wheat cultivars for sustainable agriculture in Saudi Arabia to meet the needs of the rapidly increasing populations.
Genetic deterioration in cultivated wheat provides a solid foundation for assessing genetic diversity among its relatives. This study also assess the potential for improving the efficacy of plant materials and, consequently, their utility in breeding programs, ultimately contributing to increased food production. In this investigation, we utilized a collection of 80 bread wheat germplasm provided by the United States Department of Agriculture (USDA). As a result, the primary objective of this research was to identify genetic diversity utilizing SCoT markers to facilitate sustainable production for the rapidly expanding global population.
Materials and methods
Plant material and DNA isolation
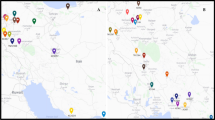
During the present study, a total of 80 bread wheat genotypes were used for molecular characterization. The plant material used during the study is given in Table 1. These genotypes were provided by the USDA and they originate from several countries and regions: South America (Argentina, Brazil, and Chile), North America (Canada), Southeast Europe (Croatia), Central Europe (Czech Republic), Eastern Europe (Hungary), Australia, East Asia (China), Southern Asia (India), Southern Europe (North Macedonia), Southeastern Europe (Serbia), Western Asia (Turkey), Eastern Europe (Romania, Ukraine), Eastern Europe and North Asia (Russia), Southwest Asia (Saudi Arabia, Yemen), and North America (United States of America, USA). To isolate the genomic DNA, all bread wheat accessions were seeded in the greenhouse, and the newly sprouted and soft leaves of those plants were collected. The DNA extraction was done using the Cetyl trimethylammonium bromide (CTAB) protocol (Doyle and Doyle 1990), with a specific protocol recommended by Diversity Arrays Technology (Nadeem et al. 2021). The quantification of isolated DNA samples was conducted utilizing a 0.8% agarose gel and the NanoDrop instrument (DS11 FX, DeNovix Inc., Wilmington, DE, USA). A final concentration of 5 ng/μL was prepared in order to facilitate subsequent polymerase chain reaction (PCR) analysis.
PCR amplification using SCoT primers
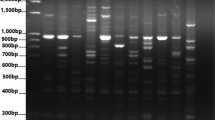
During the preliminary phase of the investigation, a random selection of six DNA samples was done and used in order to identify polymorphic markers suitable for wheat. These selected materials were then subjected to evaluation using a set of 30 SCoT primers in order to obtain polymorphism bands and the 14 highly polymorphic primers were selected (Table 2). The PCR was performed in a 10 µl reaction mixture containing 25–30 ng DNA, 1 ul of 10 × PCR buffer containing + KCl and -MgCl2, 1.5 of o.5 µM of each SCoT primer, 1 ul of 0.2 mM dNTPs, 0.15 ul of Taq DNA polymerase (DreamTaq, Thermoscientific), 1 ul of 25 mM of DNTPs, 1 ul of MgCl2, 2.5 ul of DNA (5 ng/ul) and adjust the final volume 10 ul by adding 2.85 ul of distal water. The PCR profile included initial denaturation at 95 °C for 5 min, furthermore the PCR was programmed as follows: amplification for 35 cycles with denaturation at 95 °C for 1 min, annealing at 50 °C for 1 min, after amplification, a final extension step of 72 °C for 8 min was performed. The PCR products were separated by electrophoresis through 2% agarose gel in 1 × TBE buffer for 2 h and 30 min at 110 V. GeneRuler DNA Ladder (100–3000 bp) Mix (Thermo Scientific, SM0333) was used to identify band size.
Statistical analysis
The PCR products were scored using a binary approach, where a value of 1 indicated the presence of a band, while a value of 0 indicated its absence. The study examined several diversity measures, including gene diversity (He), Shannon's information index (I), and effective alleles number (Ne), using Popgene ver. 1.32 software (Yeh et al. 2000). The Nei’s genetic distance among 80 bread wheat genotypes was also calculated using Popgene ver. 1.32 software. The polymorphism information contents (PIC) was calculated using the formula derived by Roldán-Ruiz et al. (2000) as PIC = 2fi (1—fi). The variable "fi" denotes the frequency of loci that are now present in a molecular marker, whereas the quantity (1 − fi) reflects the frequency of loci that are absent. Principal coordinate analysis (PCoA) and analysis of molecular variance (AMOVA) were performed using the GenAlExV6.5 program. In order to investigate the genetic relatedness between 80 bread wheat genotypes, a neighbor-joining analysis was conducted utilizing the R statistical software. The software known as STRUCTURE was employed in the investigation of the population structure of bread wheat germplasm, as described by Pritchard et al. (2000). The determination of the optimal number of clusters (K subpopulations) was performed by conducting the analysis three times, using the methodology described in the study conducted by Evanno et al. (2005). The range of clusters considered was from 1 to 10. For each run, the burn-in and Markov Chain Monte Carlo (MCMC) were both set to 50,000, and the number of iterations was set to 10. Subsequently, the evaluated results were subjected to processing using STRUCTURE HARVESTER v.0.9.94 (Earl 2012) in order to examine the optimal K value. The R package "pophelper" was utilized to visually represent the ΔK value that was deemed most desirable (Francis 2017).
Results
In this investigation, a total of 14 Start Codon Targeted (SCoT) primers were utilized for the purpose of assessing the genetic diversity and molecular characterization of wheat germplasm collected from various regions of the world. The 14 SCoT primers produced a total of 399 bands with an average of 28.05 bands per primer (Table 3). The maximum number of bands was produced by the primers SCoT20 (35 bands), SCoT29 (34 bands), and SCoT12 (31 bands). Furthermore, primers SCoT2, SCoT27, and SCoT28 yielded 29 bands, while SCoT3, SCoT24, and SCoT30 produced 28 bands.Out of 14 SCoT primers, SCoT-18 yileded the minimum number of bands (22). Among the 399 total bands that, a majority of 329 bands, representing an average of 82.54%, exhibited a significant level of polymorphism. It is noteworthy that each primer utilized in the study had a polymorphic band frequency of 23.5%. In addition, none of the single SCoT primers showed 100% polymorphism, and the maximum number of polymorphic bands, 28 of 29, were produced by SCoT2 with an average of 96.55%, followed by SCoT12, which produced 29 of 31 bands with an average of 93.55%, and SCoT24, which produced 26 of 28 bands with an average of 92.86%. The minimum number of polymorphisms was recorded in SCoT13, which produces 17 of 27 bands with an average of 62.96%. Furthermore, the PIC value ranged between 0.59 for the SCoT23 and 0.26 for the SCoT30, with an average PIC value of 0.39 for all tested primers. The maximum number of effective alleles (ne) of 1.59 was observed with SCoT23, followed by 1.52 with SCoT2, and the minimum ne of 1.27 was recorded with SCoT24. The average ne of 1.39 was recorded for all utilized SCoT primers. Moreover, the maximum and minimum Shannon’s information index (I) of 0.30 and 0.51 were recorded for SCoT27 and SCoT23, respectively, and the average I for all SCoT primers was 0.38. The maximum gene diversity (He) of 0.34 with ScoT23 and minimum of 0.18 with SCoT27 were recorded, and a mean gene diversity value of 0.24 was observed for all SCoT tested primers. During the evaluation of the final consequences, a mean genetic distance of 0.242 was observed, with a minimum genetic distance of 0.08 observed between Saudiarabia-1 and Saudiarabia-5, and a high genetic distance of 0.432 observed between Chile-3 and India-4. Results of the analysis of molecular variance (AMOVA) concluded that a higher number of genetic variations (92%) are present within the assessing population as compared among the populations (8%) (Table 4).
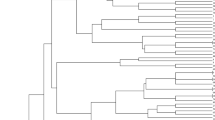
The model-based structure algorithm was performed to check the genetic variation of all tested populations and results revealed that the 80 wheat accessions were divided into two populations (population A and population B) on the basis of their collection regions, while one accession was unclassified (Fig. 1). Furthermore, population A contained a total of 40 wheat accessions; on the other hand, 39 wheat accessions were present in population B with one unclassified population (India 4). Moreover, the mean fixation index was found 0.252 and 0.25 in population A and B respectively. The Neighbor-joining analysis separated the whole germplasm of wheat into three groups (A, B & C) on the basis of their collection points (Fig. 2). The principal coordinate analysis (PCoA) strengthen the clustering of model-based structure algorithm by separating the 80 collected wheat accessions into two populations and results is clearly showed that the population A has more genetic diversity then population B (Fig. 3).
Discussion
Molecular markers are widely observed as the most reliable tools for various genetic research applications, encompassing the characterization of genetic polymorphism at the DNA level, estimation of genetic distance, determination of genetic parentage, gene mapping, analysis of genetic variation, and marker-assisted selection (Nadeem et al. 2018). Several distinct molecular marker technologies have been developed, each based on a slightly different set of guiding principles and methods. Investigation of diversity genetic of bread wheat genotypes is an effective strategy to identify unique populations with specific applications for use in short term and long term purposes of plant breeding (Mhlaba et al. 2018). Therefore, a number of molecular markers such as simple sequence repeats (SSR) (Ahmed et al. 2020), random amplified polymorphic DNA (RAPD) (Nazarzadeh et al. 2020), inter-simple sequence repeats (ISSRs) (Nosair 2021), start codon targeted (SCoT) (Nouri et al. 2021), amplified fragment length polymorphisms (AFLPs) (Roncallo et al. 2019), iPBS-retrotransposons (Nadeem 2021) and sequence related amplified polymorphism (SRAP) (Khaled et al. 2021) marker are well-known and have been explored for their potential use in wheat to assess genetic diversity and their characterizations. Mohammadi et al. (2022) conducted an investigation aimed at exploring the correlation between SCoT markers and agro-physiological traits in durum wheat breeding germplasm. A total of 220 durum wheat genotypes, acquired from ICARDA to CIMMYT, underwent evaluation for agronomic, phenological, and physiological traits over two consecutive cropping seasons (2017–18 and 2018–19). Genotyping was performed using five SCoT markers, and a regression analysis method was employed to identify informative markers associated with each trait. Results revealed significant variation in agro-physiological traits among the studied germplasms. ICARDA germplasms, favoring traits like grain yield, 1000-kernel weight, normalized difference vegetation index, plant height, and peduncle length, exhibited distinctions from CIMMYT germplasms, emphasizing the potential for drought tolerance in ICARDA-derived lines. Thirteen SCoT loci demonstrated associations with at least three traits, enhancing parental selection efficiency. Six repeatable SCoT loci emerged as candidate markers for broader germplasm assessments and genome scanning. This study, utilizing phenotypic data and SCoT markers, uncovers substantial diversity in durum wheat breeding germplasm, providing valuable insights for breeding programs, including germplasm grouping, high-yielding genotype development, and cross-breeding initiatives.
During the present study, a total of 14 SCoT primers were utilized to evaluate the genetic diversity and molecular characterization of wheat germplasms. The applied 14 SCoT primers produced a total of 399 bands with an average of 28.05 bands per primer. Total and polymorphic bands reported in this study were higher than in previous research (Pour-Aboughadareh et al. 2017; Mohamed et al. 2017; Ibrahim et al. 2017; Gholamian et al. 2019; Abouseada et al. 2023). Abdel-Lateif and Hewedy (2018) utilized a total of six SCoT primers to assess genetic diversity among eight Egyptian wheat cultivars. They reported that each primer produced an average of 3.6 bands. In another study by Kheibari et al. (2020), SCoT markers were used for the assessment of rainfed durum wheat and the average number of bands was (12.07), which was lower than our study. Alshehri (2019) investigated genetic diversity among different wheat cultivars using SCoT markers, and the average number of bands found in their study was 5.6, while an average of 7.8 bands was found in the study of Shaban et al. (2022), who also used SCoT markers. These results were lower than in our present study. Etminan et al. (2016) found an average of 9 bands using SCoT markers during the assessment of genetic diversity in wheat germplasm. The polymorphism (%) ranged from 62.96 to 96.55% with an average value of 82.54%, which were higher than that reported by Nazarzadeh et al. (2020) using RAPD and ISSR markers, Kumar et al. (2020) using ISSR marker Alshehri et al. (2020), using SCoT and ISSR primers, Mohamed et al. (2017) using SCoT primers, Çifçi and Yağdi (2012) using RAPD markers and Abouseada et al. (2023) using SCoT and ISSR primers. Etminan et al. (2016) found 100% polymorphism using the SCoT markers during the assessment of genetic diversity in wheat germplasm. Nosair (2021) reported 98.57% polymorphism in seven Egyptian wheat (Triticum aestivum L) cultivars using Scot markers.
The PIC is a frequently used metric for determining the utility of molecular markers (Houmanat et al. 2016). In the present study, the maximum PIC value recorded was 0.59 for the SCoT23, while the minimum value was 0.26 for the SCoT30. The average PIC value for all tested primers was 0.39. The PIC value of the present investigation (0.39) was in line with Etminan et al. (2016), who also used SCoT markers for the assessment of genetic variability among durum wheat. The mean PIC value reported here were found to be higher than earlier research by Mohamed et al. (2017) and Abouseada et al. (2023). They used SCoT markers for the assessment of wheat germplasm. Ibrahim et al. (2017) also used SCoT markers and revealed a PIC value of 0.24 during the assessment of genetic diversity in wheat cultivars. Gholamian et al. (2019) and Alemu et al. (2020) reported lower PIC values than our research. The higher PIC value showed the effectiveness and utility of SCoT primers for assessing the genetic diversity in wheat germplasm (Yıldız et al. 2022). The highest effective alleles (ne), 1.59, was recorded with SCoT23, followed by SCoT2, while the average ne of 1.39 was recorded for all tested SCoT primers. A greater number of effective alleles is favorable as it indicates a higher level of genetic variability. The effective number of alleles (ne) reported in this study was greater than in previous wheat research (Tajibayev et al. 2023; Nadeem et al. 2021; Marzang et al. 2020) Shannon’s information index is an important criterion for recognizing the variation, as it differentiates the genetic diversity in a population by combining abundance and evenness (Ali et al. 2019). The average Shannon’s information index (I) for SCoT primers observed at 0.38 was higher than the previous wheat research of different marker systems (Nadeem et al. 2021; Marzang et al. 2020) The result of I was in line with Tajibayev et al. (2023), who used iPBS-retrotransposons markers for the assessment of genetic variation in spring durum wheat. The maximum gene diversity (h) of 0.34 was observed with ScoT23, and the minimum of 0.18 was recorded with SCoT27. The mean h value of 0.24 was observed for all testedSCoTprimers in wheat diversity, which was greater than Nadeem et al. (2021), who reported 0.21 and 0.20 for landraces and cultivars, respectively. Tajibayev et al. (2023) reported gene diversity of 0.25, which was approximately equal to the gene diversity value of our present study. The average genetic distance was observed to be 0.242 in the present investigation, with the minimum genetic distance of 0.08 (Saudiarabia-1 and Saudiarabia-5) and the maximum genetic distance of 0.432 (Chile-3 and India-4). Results from the Analysis of Molecular Variance (AMOVA) concluded that a higher proportion of genetic variations (92%) exists within the assessing population compared to among the populations (8%) (Table 4). These AMOVA results align with findings by Etminan et al. (2016), Mourad et al. (2020) and Darvıshzadeh and Bernousı (2012). The results of AMOVA were greater than in other wheat research (Nadeem et al. 2021; Tajibayev et al. 2023).
The model-based structure algorithm grouped 80 bread wheat genotypes into two populations (Population A and B) and one unclassified population (Hungary-2) based on a membership coefficient = > 75% (Fig. 1). Population A was found to be almost equal to population B, accounting for 50% (40 genotypes). Population B comprised a total of 39 wheat genotypes, representing 48.75% of all grouped genotypes. Only Population A consisted of accession numbers collected from South America (Argentina, Brazil, and Chile), North America (Canada), Southeast Europe (Croatia), Central Europe (Czech Republic), Eastern Europe (Hungary), Australia, and East Asia (China). Population B consisted of the accession collected from Southern Asia (India), Southern Europe (North Macedonia), Southeastern Europe (Serbia), Southeastern Europe and Western Asia (Turkey), Eastern Europe (Romania, Ukraine), Eastern Europe and North Asia (Russia), Southwest Asia (Saudi Arabia, Yemen), and North America (United States of America, USA). The results of the structure analysis indicate that accessions from the same regions or their neighboring regions demonstrated similarity with each other. For example, in population A, accessions collected from South America showed genetic similarity with North American accessions, while similarity was observed between accessions from Southeast Europe and Central Europe with those from Eastern Europe. A similar level of genetic similarity was also observed in population B and it was confirmed through fixation index resulted from structure analysis. The fixation index (FST) measures the degree of gene divergence between populations based on allele frequencies. A low level of genetic differentiation is indicated by an FST of less than 0.05, whereas a moderate level of genetic differentiation is indicated by an FST of 0.05 to 0.15. Furthermore, a considerable genetic differentiation is indicated by an FST of 0.15 to 0.25, and a very large genetic differentiation is indicated by an FST greater than 0.25 (Meirmans and Hedrick, 2011). In this study, we observed FST value of 0.252 and 025 for population A and B respectively and these values confirmed the presence of moderate to large level of genetic differentiation between both populations. The neighbor-joining dendrogram analysis divided the examined wheat germplasm into three groups (Groups A, B, and C), primarily distinguished by the locations of their respective collection points (Fig. 2). Groups A and B are well-defined such as group A contained of 39 accessions while group B was consisted on 40 accessions of wheat but group C consists only of one accession (India 4). There are various reasons why this single accession is placed in a separate group. It could be due to impurities in its DNA, or significant novelties that differentiate it from all others. This separation was also observed in our model-based structure algorithm, where it was placed in an unclassified population. With few exceptions, the Neighbor-joining clustering method exhibited a clustering pattern similar to the model-based structure, dividing the entire germplasm into two to three groups. Notably, accessions from Chilli, Australia, and Brazil were consistently grouped together, mirroring the patterns observed in population structures. Although certain accessions were placed in-group B in the Neighbor-joining tree, their presence in-group A was evident in the population structure. Despite this discrepancy, these accessions clustered together in the Neighbor-joining method. Notably, accessions from the same countries consistently grouped together, either among themselves or with accessions from other countries. The clustering of accessions from different regions implies shared genetic similarities (Yildiz et al. 2022). The structure clustering was preferred over other clustering algorithms as it has been demonstrated to be more reliable and significantly more illuminating (Newell et al. 2013; Bouchet et al. 2012). In addition, different diversity parameters were computed for the populations that underwent the structure evaluation, revealing the presence of greater genetic variations. This was further confirmed by the AMOVA study for the structure-evaluated population that there is more genetic variation within populations than among them. The aggregation of model-based structure was validated by the PCoA analysis, which also contributed to the division of the wheat germplasm into two distinct populations (Fig. 3). Hence, it can be argued that the evaluated wheat germplasm possesses a substantial amount of genetic variations within the population (92%), potentially beneficial for future crop breeding efforts.
Conclusion
In conclusion, research employing SCoT markers has revealed significant genetic variability within tested wheat germplasm, presenting great potential for safeguarding forthcoming food sustainability. The investigation of a wide range of genetic resources presents a valuable collection of prospective characteristics, allowing breeders to cultivate enhanced wheat cultivars that possess increased adaptability, disease resistance, and higher yields. Genetic diversity plays a crucial role in mitigating global concerns, including climate change and population expansion, which pose significant threats to food production and sustainability. Through the utilization of the valuable data obtained from SCoT markers, scholars and decision-makers can make substantial advancements in strengthening food security, ultimately ensuring a more resilient and productive agricultural future.
Availability of data and materials
All data about this study are present within the manuscript.
References
Abd El-Moneim D, Alqahtani MM, Abdein MA, Germoush MO (2020) Drought and salinity stress response in wheat: physiological and TaNAC gene expression analysis in contrasting Egyptian wheat genotypes. J Plant Biotech 47(1):1–14
Abdel-Lateif KS, Hewedy OA (2018) Genetic diversity among Egyptian wheat cultivars using SCoT and ISSR markers. Sabrao J Breed Genet 50:36–45
Abouseada HH, Mohamed AS, Teleb SS, Badr A, Tantawy ME, Ibrahim SD, Ellmouni FY, Ibrahim M (2023) Genetic diversity analysis in wheat cultivars using SCoT and ISSR markers, chloroplast DNA barcoding and grain SEM. BMC Plant Biol 23:1–5
Ahmed HG, Kashif M, Rashid MA, Sajjad M, Zeng Y (2020) Genome wide diversity in bread wheat evaluated by SSR markers. Intl J Agric Biol 24:263–272
Alemu A, Feyissa T, Letta T, Abeyo B (2020) Genetic diversity and population structure analysis based on the high-density SNP markers in Ethiopian durum wheat (Triticum turgidum ssp durum). BMC Gen 21(1):1–12
Ali F, Yılmaz A, Nadeem MA, Habyarimana E, Subaşı I, Nawaz MA, Baloch FS (2019) Mobile genomic element diversity in world collection of safflower (Carthamus tinctorius L) panel using iPBS-retrotransposon markers. PLoS ONE 14(2):e0211985
Al-Qurainy F, Khan S, Nadeem M, Tarroum M (2015) SCoT marker for the assessment of genetic diversity in Saudi Arabian date palm cultivars. Paki J Bot 47(2):637–643
Alshehri MA (2019) Genetic variation of Saudi wheat genotypes through ISSR and SCoT assays. Int J Pharm Phytopharm Res 9:57–65
Alshehri MA, Alzahrani O, Aziza AT, Alasmari A, Ibrahim S, Bahattab O, Alduaydi SA (2020) Correlation and genetic analyses of different characteristics in Saudi Arabian wheat reveal correlation networks and several traitassociated markers. JAPS. https://doi.org/10.36899/JAPS.2020.6.0169
Baloch FS, Nadeem MA (2022) Unlocking the genomic regions associated with seed protein contents in Turkishcommon bean germplasm through genome-wide association study. Turk J Agri Forest 46(1):113–128
Baloch FS, Guizado SJ, Altaf MT, Yüce I, Çilesiz Y, Bedir M, Nadeem MA, Hatipoglu R, Gómez JC (2022) Applicability of inter-primer binding site iPBS-retrotransposon marker system for the assessment of genetic diversity and population structure of Peruvian rosewood (Aniba rosaeodora Ducke) germplasm. Mol Biol Rep 49(4):2553–2564
Battersby J (2017) Food system transformation in the absence of food system planning: the case of supermarket and shopping mall retail expansion in Cape Town. South Africa Built Envirn 43(3):417–430
Bouchet S, Pot D, Deu M, Rami JF, Billot C, Perrier X, Rivallan R, Gardes L, Xia L, Wenzl P, Kilian A (2012) Genetic structure, linkage disequilibrium and signature of selection in sorghum: lessons from physically anchored DArT markers. PLoS ONE 7:e33470
Braun HJ, Atlin G, Payne T (2010) Multi-location testing as a tool to identify plant response to global climate change. Cli Chan Crop Prod 1:115–138
Çifçi EA, Yağdi K (2012) Study of genetic diversity in wheat (Triticum aestıvum) varities using random amplified polymorphic DNA (RAPD) analysis. Turk J Field Crop 17:91–95
Collard BC, Mackill DJ (2009) Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Boil Rep 27:86–93
Darvishzadeh R, Bernousi I (2012) Molecular similarity relationships among Iranian bread wheat cultivars and breeding lines using ISSR markers. Notul Bot Horti Agrob Cluj-Nap 40(2):254–260
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:39–40
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
El-Moneim A (2020) Characterization of ISSR and SCoT markers and TaWRKY gene expression in some Egyptian wheat genotypes under drought stress. J Plant Prod Sci 8:31–46
Etminan A, Pour-Aboughadareh A, Mohammadi R, Ahmadi-Rad A, Noori A, Mahdavian Z, Moradi Z (2016) Applicability of start codon targeted (SCoT) and inter-simple sequence repeat (ISSR) markers for genetic diversity analysis in durum wheat genotypes. Biotechnol Biotechnol Equip 30:1075–1081
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
FAO Ifad & WFP (2015) The state of food insecurity in the world 2015. Meeting the 2015 international hunger targets: taking stock of uneven progress. FAO, Rome
FAO 2008 Opportunities and challenges of biofuel production for food security and the environment in Latin America and the Caribbean. Document LARC/8/4 for the 30th Session of the FAO Regional Conference for Latin America and the Caribbean, Brasilia, Brazil, 14–18 April 2008. http://www.fao.org/Unfao/Bodies/RegConferences/Larc30/Index_en.htm
Francis RM (2017) pophelper: an R package and web app to analyse and visualize population structure. Mol Ecol Resour 2017:27–32
Gholamian F, Etminan A, Changizi M, Khaghani S, Gomarian M (2019) Assessment of genetic diversity in Triticum urartu Thumanjan ex Gandilyan accessions using start codon targeted polymorphism (SCoT) and CAAT-box derived polymorphism (CBDP) markers. Biotechnol Biotechnol Equip 33:1653–1662
Guo DL, Zhang JY, Liu CH (2012) Genetic diversity in some grape varieties revealed by SCoT analyses. Mol Biol Rep 39:5307–5313
Hamidi H, Talebi R, Keshavarzi F (2014) Comparative efficiency of functional gene-based markers, start codon targeted polymorphism (SCoT) and conserved DNA-derived polymorphism (CDDP) with ISSR markers for diagnostic fingerprinting in wheat (Triticum aestivum L). Cereal Research Communications 42(4):558–567
Hegde SG, Valkoun J, Waines JG (2002) Genetic diversity in wild and weedy Aegilops, Amblyopyrum, and Secale species—a preliminary survey. Crop Sci 42:608–614
Houmanat K, Charafi J, Mazouz H, El Fechtali M, Nabloussi A (2016) Genetic diversity analysis of safflower (Carthamus tinctorius L.) accessions from different geographic origins using ISSR markers. Int J Agric Biol 18:881–887
Ibrahim M, Mohamed AS, Teleb S, Ibrahim S, Tantawy M (2017) Taxonomic and molecular study on some Asian cultivars of Triticum aestivum L. Taeckholmia 37:16–29
Khaled AG, Elameen TM, Ahmed AY, Mohiy M, Elshazly IF (2021) SRAP molecular markers linked to three morphological traits in Egyptian bread wheat (Triticum aestivum L.). SVU-Int J Agric Sci 3:145–158
Kumar P, Sharma V, Sanger RS, Kumar P, Yadav MK (2020) Analysis of molecular variation among diverse background wheat (Triticum aestivum L) genotypes with the help of ISSR markers. IJCS 8(3):271–276
Leegood RC, Evans JR, Furbank RT (2010) Food security requires genetic advances to increase farm yields. Nature 464(7290):831–831
Leśniowska-Nowak J, Sozoniuk M, Kawęcka M, Maga KO, Mrozek MA (2020) Identification of Triticum durum genotypes showing increased tolerance to oxidative stress. Agronomy Sci 75(1):43–65
Leśniowska-Nowak J, Okoń S, Wieremczuk A (2021) Molecular diversity analysis of genotypes from four Aegilops species based on retrotransposon–microsatellite amplified polymorphism (REMAP) markers. Cereal Res Commun 49:37–44
Mafakheri M, Kordrostami M, Rahimi M, Matthews PD (2020) Evaluating genetic diversity and structure of a wild hop (Humulus lupulus L.) germplasm using morphological and molecular characteristics. Euphytica 216:1–9
Marzang N, AbdollahiMandoulakani B, Shaaf S, Ghadimzadeh M, Bernousi I, AbbasiHolasou H, Sadeghzadeh B (2020) IRAP and REMAP-based genetic diversity among Iranian, Turkish, and International Durum wheat (Triticum turgidum L) cultivars. J Agric Sci Technol 22(1):271–285
Meirmans PG, Hedrick PW (2011) Assessing population structure: FST and related measures. Mol Ecol Res 11(1):5–18
Mhlaba ZB, Mashilo J, Shimelis H, Assefa AB, Modi AT (2018) Progress in genetic analysis and breeding of tepary bean (Phaseolus acutifolius A. Gray): A review. Sci Hortic 237:112–119
Mohamed AS, Ibrahim M, Teleb SS, Tantawy ME (2017) SEM and SCoT markers unveil new taxonomic and genetic insights about some Northern African Triticum aestivum L. Cultivars Vegetos 30:1. https://doi.org/10.4172/2229.2017;4473
Mohammadi R, Cheghamirza K, Geravandi M, Abbasi S (2022) Assessment of genetic and agro-physiological diversity in a global durum wheat germplasm. Cereal Res Commun 50(1):117–126
MoradiKheibari ZMK, Azizinezhad R, Mehrabi AM, Khosrowshahli M, Etminan A (2020) Molecular diversity and genetic structure of rainfed durum wheat genotypes using SCoT markers. Iran. J. Genet. Plant Breed 9(2):115–125
Mourad AM, Belamkar V, Baenziger PS (2020) Molecular genetic analysis of spring wheat core collection using genetic diversity, population structure, and linkage disequilibrium. BMC Genom 21:1–12
Nadeem MA (2021) Deciphering the genetic diversity and population structure of Turkish bread wheat germplasm using iPBS-retrotransposons markers. Mol Biol Rep 48:6739–6748
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H (2018) DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip 32:261–285
Nazarzadeh Z, Onsori H, Akrami S (2020) Genetic diversity of bread wheat (Triticum aestivum L.) genotypes using RAPD and ISSR molecular markers. J Genet Resou 6:69–76
Newell MA, Cook D, Hofmann H, Jannink JL (2013) An algorithm for deciding the number of clusters and validation using simulated data with application to exploring crop population structure. Ann App Stat 1:1898–1916
Nosair HR (2021) Genetic diversity studies on seven Egyptian wheat (Triticum aestivum) cultivars using SCoT and ISSR polymorphism markers. Taeckholmia 40:143–151
Nouri A, Golabadi M, Etminan A, Rezaei A, Mehrabi AA (2021) Comparative assessment of SCoT and ISSR markers for analysis of genetic diversity and population structure in some Aegilops tauschii Coss. accessions. Plant Genet Resou 19:375–383
Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Etminan A, Moghaddam M, Siddique KH (2017) Physiological responses to drought stress in wild relatives of wheat: implications for wheat improvement. Acta Physiol Plant 39(4):106
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Roldán-Ruiz I, Dendauw J, Van Bockstaele E, Depicker A, De Loose M (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6(2):125–134. https://doi.org/10.1023/A:1009680614564
Roncallo PF, Beaufort V, Larsen AO, Dreisigacker S, Echenique V (2019) Genetic diversity and linkage disequilibrium using SNP (KASP) and AFLP markers in a worldwide durum wheat (Triticum turgidum L. var durum) collection. PLoS ONE 14:e0218562
Salama YE, Saad KI (2022) Evaluation of genetic variability of bread wheat genotypes under normal irrigation and water stress conditions using multivariate analysis. J Plant Prod 11:833–838
Shaban AS, Arab SA, Basuoni MM, Abozahra MS, Abdelkawy AM, Mohamed MM (2022) SCoT, ISSR, and SDS-PAGE investigation of genetic diversity in several egyptian wheat genotypes under normal and drought conditions. Int J Agron. https://doi.org/10.1155/2022/7024028
Tabasi M, Sheidai M, Hassani D, Koohdar F (2020) DNA fingerprinting and genetic diversity analysis with SCoT markers of Persian walnut populations (Juglans regia L.) in Iran. Genet Res Crop Evol 67:1437–1447
Tabasum A, Hameed A, Asghar MJ (2020) Exploring the genetic divergence in mungbean (Vigna radiata L.) germplasm using multiple molecular marker systems. Mol Biotechnol 62:547–556
Tajibayev D, Mukin K, Babkenov A, Chudinov V, Dababat AA, Jiyenbayeva K (2023) Exploring the agronomic performance and molecular characterization of diverse spring durum wheat germplasm in Kazakhstan. Agronomy 13(7):1955
Velu G, Crespo Herrera L, Guzman C et al (2019) Assessing genetic diversity to breed competitive biofortified wheat with enhanced grain Zn and Fe concentrations. Front Plant Sci 9:413924
WFP (2020). COVID-19 and the 5 major threats it poses to global food security. Available online at: https://www.wfpusa.org/articles/covid-19-and-global-food-security/
Xiong F, Zhong R, Han Z, He L, Zhuang W, Tang R (2011) Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol Biol Rep 38:3487–3494
Yeh FC, Yang R, Boyle TJ, Ye Z, Xiyan JM (2000) PopGene32, Microsoft Windows-based freeware for population genetic analysis, version 1.32. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton
Yalinkiliç NA, Baþbað S, Altaf MT, Ali A, Nadeem MA, Baloch FS (2024) Applicability of SCoT markers in unraveling genetic variation and population structure among sugar beet (Beta vulgaris L.) germplasm. Mol Biol Rep 51(1):584
Yıldız M, Altaf M, Baloch F, Koçak M, Sadik G, Kuzgun C, Nadeem M, Ali F, Bedir M, Tunçtürk M (2022) Assessment of genetic diversity among 131 safflower (Carthamus tinctorius L.) accessions using peroxidase gene polymorphism (POGP) markers. Mol Biol Rep 49(7):6531–6539
Yilmaz A, Ciftci V (2021) Genetic relationships and diversity analysis in Turkish laurel (Laurus nobilis L.) germplasm using ISSR and SCoT markers. Mol Biol Rep 48(5):4537–4547
Zarei A, Erfani-Moghadam J (2021) SCoT markers provide insight into the genetic diversity, population structure and phylogenetic relationships among three Pistacia species of Iran. Genet Res Crop Evol 68(4):1625–1643
Zhao C, Liu B, Piao S, Wang X (2017) Temperature increase reduces global yields of major crops in four independent estimates. Proceed Nat Acad Sci 114(35):9326–9331
Acknowledgements
The authors are thankful to USDA for providing plant material for this study.
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK).
Author information
Authors and Affiliations
Contributions
Conceptualization: MAN, Supervision: MAN, FSB, Experimentation: MAN, AA, MTA, NB, WL, MB, Data Curation: AA, NB, MTA, WL, MB, Statistical analysis: MAN, Validation: MAN, FSB, First draft: AA, MKI, Review, and editing: AI, AG, Manuscript revision: MA, HD, MAN.
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. All authors have read and shown their willingness to publish this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Altaf, M.T., Nadeem, M.A., Ali, A. et al. Applicability of Start Codon Targeted (SCoT) markers for the assessment of genetic diversity in bread wheat germplasm. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-02016-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-02016-0