Abstract
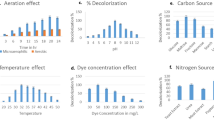
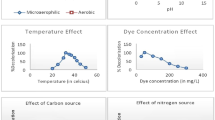
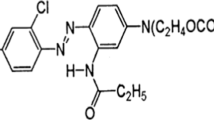
Dyes primarily present in effluents from textile industries are recalcitrant organic molecules with a complex aromatic structure, which are considered difficult to eliminate, degrade and detoxify biologically. In the current study, Pseudomonas fluorescens was used to degrade Procion Red Yellow. The bacterium P. fluorescens NCIM 2100 was procured from National Chemical Laboratory (NCL), Pune, India, that was well adapted to grow and survive in broth supplemented with dyes. The dye was exposed to bacterium followed by the identification of its degradation products by a combination of UV, 1H NMR and IR spectrophotometry. After exposure, the dye was firstly broken down into an intermediate compound lacking azo benzene group. This compound further changed into another intermediate compound that may contain either OH or NH group. This intermediate finally changed into 3, 5-diamino-1, 4, 6, 8-tetrahydroxy-2,7-naphthyldisulfonic acid sodium salt or 1, 3, 4, 5, 6-hexa-hydroxy-2, 7-naphthyldisulfonic acid sodium salt in broth. These breakdown or transformation products were innoxious in nature. Thus P. fluorescens can be used as a promising candidate for the bioremediation of the textile effluents containing Procion Red Yellow to offer promise for environmental decontamination and even greater development for green chemistry in the near future.




Similar content being viewed by others
References
Bentzien J, Hickey ER, Kemper RA, Brewer ML, Dyekjær JD, East SP, Whittaker M (2010) An in silico method for predicting Ames activities of primary aromatic amines by calculating the stabilities of nitrenium ions. J Chem Inf Model 50:274–297
Brüschweiler BJ, Merlot C (2017) Azo dyes in clothing textiles can be cleaved into a series of mutagenic aromatic amines which are not regulated yet. Regul Toxicol Pharmacol 88:214–226
Fan HJ, Huang ST, Chung WH, Jan JL, Lin WY, Chen CC (2009) Degradation pathways of crystal violet by Fenton and Fenton-like systems: condition optimization and intermediate separation and identification. J Hazard Mater 171:1032–1044
Mathur N, Bhatnagar P, Sharma P (2012) Review of the mutagenicity of textile dye products. Univers J Environ Res Technol 2:1–18
Singh K, Arora S (2011) Removal of synthetic textile dyes from wastewaters: a critical review on present treatment technologies. Crit Rev Environ Sci Technol 41:807–878
Holkar CR, Jadhav AJ, Pinjari DV, Mahamuni NM, Pandit AB (2016) A critical review on textile wastewater treatments: possible approaches. J Environ Manag 182:351–366
Pandey A, Singh P, Iyengar L (2007) Bacterial decolorization and degradation of azo dyes. Int Biodeter Biodegrad 59:73–84
Sarayu K, Sandhya S (2010) Aerobic biodegradation pathway for Remazol Orange by Pseudomonas aeruginosa. Appl Biochem Biotechnol 160:1241–1253
Lade HS, Waghmode TR, Kadam AA, Govindwar SP (2012) Enhanced biodegradation and detoxification of disperse azo dye Rubine GFL and textile industry effluent by defined fungal-bacterial consortium. Int Biodeterior Biodegrad 72:94–107
Garg SK, Tripathi M (2017) Microbial strategies for discoloration and detoxification of azo dyes from textile effluents. Res J Microbiol 12:1–19
Peng YH, Shih YH (2013) Microbial degradation of some halogenated compounds: Biochemical and molecular features. In: Chamy R, Rosenkranz F (eds) Biodegradation of hazardous and special products, Chapter 4. InTech, Rijeka, pp 51–69
Nikolaivits E, Dimarogona M, Fokialakis N, Topakas E (2017) Marine-derived biocatalysts: importance, accessing, and application in aromatic pollutant bioremediation. Front Microbiol 8:265
Ju KS, Parales RE (2010) Nitroaromatic compounds, from synthesis to biodegradation. Microbiol Mol Biol Rev 74:250–272
Hsueha CC, Chen CT, Hsua AW, Wub CC, Chen BY (2017) Comparative assessment of azo dyes and nitroaromatic compounds reduction using indigenous dye-decolorizing bacteria. J Taiwan Inst Chem Eng 79:134–140
Tišma M, Zelić B, Vasić-Rački D (2010) White-rot fungi in phenols, dyes and other xenobiotics treatment—a brief review. Croat J Food Sci Technol 2:34–47
Magan N, Fragoeiro S, Bastos C (2010) Environmental factors and bioremediation of xenobiotics using white rot fungi. Mycobiology 38:238–248
Sugumar RW, Sadanandan S (2010) Combined anaerobic-aerobic bacterial degradation of dyes. Eur J Chem 7:739–744
Karatas M, Dursun S, Argum ME (2010) The decolorization of azo dye reactive Black 5 in a sequential anaerobic-aerobic system. Ekoloji 19:15–23
Mahmood S, Khalid A, Arshad M, Mahmood T, Crowley DE (2015) Detoxification of azo dyes by bacterial oxidoreductase enzymes. Crit Rev Biotechnol 36(4):1–13
Arora PK (2015) Bacterial degradation of monocyclic aromatic amines. Front Microbiol 6:820
Agrawal S, Tipre D, Patel B, Dave S (2017) Bacterial decolourization, degradation and detoxification of Azo dyes: An eco-friendly approach. In: Kalia V, Kumar P (eds) Microbial applications, vol 1. Springer, Cham
Ghodake G, Jadhav U, Tamboli D, Kagalkar A, Govindwar S (2011) Decolorization of textile dyes and degradation of mono-azo dye Amaranth by Acinetobacter calcoaceticus NCIM 2890. Indian J Microbiol 51:501–508
Rani B, Kumar V, Singh J, Bisht S, Teotia P, Sharma S, Kela R (2014) Bioremediation of dyes by fungi isolated from contaminated dye effluent sites for bio-usability. Braz J Microbiol 45:1055–1063
Pandey BV, Upadhyay RS (2006) Spectroscopic characterization and identification of Pseudomonas fluorescens mediated metabolic product of Acid Yellow-9. Microbiol Res 161:311–315
Pandey BV, Upadhyay RS (2010) Pseudomonas fluorescens can be used for bioremediation of textile effluents Direct Orange -102. Trop Ecol 51:397–403
Pandey RR, Pandey BV, Upadhyay RS (2011) Characterization and identification of Pseudomonas fluorescens NCIM 2100 degraded metabolic product of Crystal Violet. Afr J Biotechnol 10:624–631
Dyer JR (1991) Application of absorption spectroscopy on organic compounds. Prentice Hall, Atlanta
Buckingham J, Macdonald F (1996) Dictionary of organic compounds, 6th edn. Electronic Publishing Division, New York
Shah MP, Patel KA, Nair SS, Darji AM (2013) Microbial decolorization of Methyl Orange dye by Pseudomonas spp. ETL-M Int J Environ Bioremediat Biodegrad 1:54–59
Shah MP, Patel KA, Nair SS, Darji AM (2013) Microbial degradation of textile dye (Remazol Black B) by Bacillus spp. ETL-2012. J Bioremed Biodeg 4:180
Acknowledgements
The authors would like to thank Dr. Rahul Singh, Department of Chemistry, B.H.U., Varanasi for giving advices on this article. The authors would further like to thank the anonymous reviewers for their helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest to publish this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance Statement
The dye Procion Red Yellow was exposed to Pseudomonas fluorescens followed by the identification of its degradation products. The breakdown products were innoxious in nature. Thus, P. fluorescens can be used as a promising candidate for the bioremediation purposes.
Rights and permissions
About this article
Cite this article
Pandey, B.V., Dubey, M.K. & Upadhyay, R.S. Bioremediation of Textile Dye Procion Red Yellow by Using Pseudomonas fluorescens. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 90, 135–141 (2020). https://doi.org/10.1007/s40011-019-01089-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-019-01089-7