Abstract
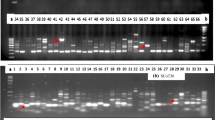
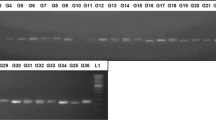
Chrysanthemum is ranked second among the most economically important cut flowers after rose. This flower is considered very significant from different aspects of commercial herbs and traditional medicine. In this study, 30 simple sequence repeat (SSR) and 8 start codon targeted polymorphism (SCoT) markers were used to assess the genetic diversity of 32 cultivated varieties of C. morifolium. In total, 127 alleles were detected. The average number of alleles per marker locus was 2.13 for SSR (ranging from 1 to 4) and 7.78 for SCoT markers (ranging from 1 to 18). A positive significant correlation between similarity matrices was obtained by the two markers. Average PIC values for SSR and SCoT markers were 0.37 and 0.34, respectively. The results of clustering for both marker systems divided the genotypes into four clusters. These findings revealed that the efficiency of SSR and SCoT markers was relatively the same in fingerprinting of chrysanthemum genotypes. The authors performed combination of SCoT and SSR markers as a novel method to assess genetic diversity in chrysanthemum. Further implications of the results could be used in germplasm conservation and enhancing the genetic base of the breeding programs of C. morifolium.



Similar content being viewed by others
References
Bhattacharya A, Teixeira JA (2006) Molecular systematics in Chrysanthemum  Grandiflorum (Ramat.) Kitamura. Sci Hortic 109:379–384. https://doi.org/10.1016/j.scienta.2006.06.004
Kafi M, Ghahsareh M (2009) Floriculture, vol 1, 4th edn. Jahad Press, Tehran, pp 108–118
Al-qurainy F, Salim K, Mohammad N, Mohamed T (2015) SCoT marker for the assessment of genetic diversity in Saudi Arabian date palm cultivars. Pak J Bot 47(2):637–643
Anderson NO (2006) Flower breeding and genetics. Issues, challenges and opportunities for the 21st Century. Springer, Dordrecht
Liu P-L, Wan Q, Guo Y-P, Yang J, Rao G-Y (2012) Phylogeny of the genus Chrysanthemum L.: evidence from single-copy nuclear gene and chloroplast DNA sequences. PLoS ONE 7(11):e48970. https://doi.org/10.1371/journal.pone.0048970
Hartl DL, Jones EW (2005) Genetics: analysis of genes and genomes. Jones and Bartlett Publishers, Sudbury
Klie M, Ina M, Marcus L, Thomas D (2013) Lack of structure in the gene pool of the highly polyploid ornamental chrysanthemum. Mol Breeding 32:339. https://doi.org/10.1007/s11032-013-9874-4
Kalendar R, Flavell AJ, Ellis THN, Sjakste T, Moisy C, Schulman AH (2011) Analysis of plant diversity with retrotransposon-based molecular markers. Heredity 106:520–530
Hajibarat Z, Saidi A, Hajibarat Z, Talebi R (2014) Genetic diversity and population structure analysis of landrace and improved chickpea (Cicer Arietinum) genotypes using morphological and microsatellite markers. Environ Exp Biol 12:161–166
Collard Bertrand CY, Mackill DJ (2009) Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Biol Rep 27:86–93. https://doi.org/10.1007/s11105-008-0060-5
Hajibarat Z, Saidi A, Hajibarat Z, Talebi R (2015) Characterization of genetic diversity in chickpea using SSR markers, start codon targeted polymorphism (SCoT) and conserved DNA-derived polymorphism (CDDP). Physiol Mol Biol Plants 21(3):365–373. https://doi.org/10.1007/s12298-015-0306-2
Chatterjee J, Mandal AKA, Ranade SA, Silva JATD et al (2006) Molecular systematics in Chrysanthemum grandiflorum (Ramat.) Kitamura. Sci Hortic 110:373–378
Zhang F, Chen S, Chen F, Fang W, Li F (2010) A preliminary genetic linkage map of chrysanthemum (Chrysanthemum morifolium) cultivars using RAPD, ISSR and AFLP markers. Sci Hortic 125:422–428
Li T, Guo J, Li Y, Ning H, Xia S, Zheng C (2013) Scientia horticulturae genetic diversity assessment of chrysanthemum germplasm using conserved DNA-derived polymorphism markers. Sci Hortic 162:271–277. https://doi.org/10.1016/j.scienta.2013.08.027
Saidi A, Eghbalnegad Y, Hajibarat Z (2017) Study of genetic diversity in local rose varieties (Rosa spp.) using molecular markers. Banats J Biotechnol 16:148–157
Wang Q, Zhang B, Lu G (2009) Conserved region amplification polymorphism (CoRAP), a novel marker technique for plant genotyping in Salvia miltiorrhiza. Plant Mol Biol Rep 27:139–143
Yuan W, Ye S, Du L, Li S, Miao X, Shang F (2016) Isolation and characterization of microsatellite markers for Dendranthema morifolium (Asteraceae) using next-generation sequencing. Genet Mol Res 15(4):1–7
Zhang F, Chen S, Chen F, Fang W, Deng Y, Chang Q, Liu P (2011) Genetic analysis and associated SRAP markers for flowering traits of chrysanthemum (Chrysanthemum morifolium). Euphytica 117:15–24
Zhang Y, Chen W (2013) Assessing the genetic diversity of chrysanthemum cultivars with microsatellites. J Am Soc Hortic Sci 138(6):479–486
Zhang DL, Zhang HL, Wang MX, Sun JL, Qi YW et al (2009) Genetic structure and differentiation of Oryza sativa L. in China revealed by microsatellites. Theor Appl Genet 119:1105–1117
Feng S, Renfeng H, Jiangjie L, Mengying J, Xiaoxia S (2016) Development of SSR markers and assessment of genetic diversity in medicinal Chrysanthemum morifolium cultivars. Front Genet 7:1–11. https://doi.org/10.3389/fgene.2016.00113
Xiong F, Zhong R, Han Z, Jiang J, He L, Zhuang W, Tang R (2011) Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol Biol Rep 38:3487–3494
Zhang J, Esselink G, Che D, Fougère-Danezan M, Arens P, Smulders M (2013) The diploid origins of allopolyploid rose species studied using single nucleotide polymorphism haplotypes flanking a microsatellite repeat. J Hortic Sci Biol 88:85–92
Pirttilä MA, Hirsikorpi M, Kämäräinen T, Jaakola L, Hohtola A (2001) DNA isolation methods for medicinal and aromatic plants. Int Soc Plant Mol Biol 19:273a–273f
Huson DH, Bryant D (2006) User manual for SplitsTree4 V4.13.1. Mol Biol Evol 2 (23):254–267. software available from www. splitstree.org
Rohlf FJ (2000) NTSYS-Pc numerical taxonomy and multivariate analysis system. Exeter Publications, Setauket
Khaing AA, Moe KT, Hong WJ, Park CS, Yeon KH, Park HS (2013) Phylogenetic relationships of chrysanthemums in Korea based on novel SSR markers. Genet Mol Res 12(4):5335–5347
Bhattacharyya P, Kumaria S, Kumar S, Tandon P (2013) Start codon targeted (SCoT) marker reveals genetic diversity of Dendrobium nobile Lindl. An endangered medicinal orchid species. Gene 529:21–26
Gorji AM, Poczai P, Polgar Z, Taller J (2011) Efficiency of arbitrarily amplified dominant markers (SCOT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am J Potato Res 88:226–237
Heikrujam M, Kumar J, Agrawal V (2015) Genetic diversity analysis among male and female jojoba genotypes employing gene targeted molecular markers, start codon targeted (SCoT) polymorphism and CAAT box-derived polymorphism (CBDP) markers. MetaGene 5:90–97. https://doi.org/10.1016/j.mgene.2015.06.001
Acknowledgement
The authors are grateful to the Department of Plant Sciences and Biotechnology of Shahid Beheshti University for providing financial support for this project work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest among the authors to publish this manuscript.
Additional information
Significance Statement
The study was conducted to assess the genetic diversity of 32 cultivated varieties. Two systems were used and compared together for this work for fingerprinting. The authors, for the first time, introduce SCoT and SSR as a novel system to assess chrysanthemum.
Rights and permissions
About this article
Cite this article
Kobeissi, B., Saidi, A., Kobeissi, A. et al. Applicability of SCoT and SSR Molecular Markers for Genetic Diversity Analysis in Chrysanthemum morifolium Genotypes. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 89, 1067–1077 (2019). https://doi.org/10.1007/s40011-018-1024-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-018-1024-7