Abstract
Twenty eight inter simple sequence repeat (ISSR) markers were used to determine the genotypic identification and phylogenetic relationship within 48 varieties of Indica rice including four aromatic rice grown in eastern part of India. Out of 44 upland and low land varieties, 39 varieties released by the Orissa University of Agriculture and Technology, Orissa and five released by the other National Institutes of India. A total of 28 ISSR primers were screened representing di- and tri- nucleotide repeats, out of which 12 ISSR markers were selected for varietal analysis as diagnostic markers. The number of bands per locus ranged from 4 to 8, with an average of 6. The results revealed that all the primers showed distinct polymorphism among the varieties indicating the robust nature of ISSR markers. Most of the primers showed highest polymorphic information content and resolving power. The cluster analysis indicates that all the rice genotypes are grouped into two classes in which aromatic rice varieties clustered separately from upland and lowland varieties. Principal component analysis showed the distinct differences among themselves. Based on this study, the larger range of similarity values using ISSR markers provides greater confidence for the assessment of genetic relationships among the varieties. The information obtained from the DNA profile helps to identify the variety diagnostic markers in 48 rice genotypes. This outcome of the investigation can be used in background selections during backcross breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice, Oryza sativa (2n = 24) belonging to the family Poaceae and subfamily Oryzoidea is the staple food for one-third of the world’s population and occupies almost one-fifth of the total land area covered under cereals. It is grown under diverse conditions and over wide geographical range. Most of the world’s rice is cultivated and consumed in Asia, which constitutes more than half of the global population. Approximately 11 % of the world’s arable land is planted annually to rice, and it ranks next to wheat. The world’s rice production has doubled during last 25 years, largely due to the use of improved technology such as high yielding varieties and better crop management practices [1]. Further scope of crop improvement depends on the availability, genetic diversity and variability and use of new biotechnological tools. There exists a rich diversity in rice. Except for a few earlier studies on classification of germplasm using isozymes, there have been a few studies to assess the diversity of Indian rice varieties including landraces, at the molecular level. Presently some of the varieties have gone threatened. Hence, their conservation and characterization is of utmost important. Some of the traditional varieties are not sufficiently represented to study the genetic diversity. A number of molecular markers have been used for diversity study in rice [2–4]. Inter simple sequence repeat (ISSR) is a microsatellite-based multi locus marker technique, which is simple and useful for estimating genetic diversity in several crop plants. The technique has the advantages of random amplified polymorphic DNA (RAPD) and in addition shows higher level of polymorphism, reproducibility and cost-effectiveness per polymorphism. ISSR–PCR has been used in genetic diversity studies in several crop plants. In rice, ISSR–PCR has been used for the analysis of micro satellite motif frequency and fingerprinting of varieties, determining phylogenetic relationships among Oryza species [5–9]. The usefulness of multi locus ISSR–PCR markers in determining diversity within improved varieties, landraces of rice of Orissa has not been previously investigated. The objectives of this study are: (i) to assess the extent of genetic diversity in Indian landraces of rice as compared to improved cultivars and other accessions using ISSR primers; (ii) to compare the level of polymorphism revealed by each primer, and between landraces and varieties, (iii) to develop a rational basis for the choice of informative primers using polymorphic information content, resolving power and mean genetic similarity and (iv) to identify the variety diagnostic ISSR markers for rice improvement program.
Material and Methods
Plant Materials
Forty four elite upland and lowland rice genotypes and 4 aromatic genotypes (‘Basumati’, ‘Kalajeera’, ‘Pumpudibasa’, ‘Neelabati’) were collected from the germplasm centre maintained by the Department of Plant Breeding and Genetics, Orissa University of Agriculture and Technology, Bhubaneswar for ISSR analysis. The morphological and agronomic characteristics have also been indicated in Table 1. The seeds were sown in the earthen pots and kept in the green house for germination. Leaf samples were collected and subsequently stored at −20 °C until processed.
DNA Extraction and PCR reaction
DNA was extracted as described by Doyle and Doyle [10] with minor modifications. The reaction mixture composition and conditions were empirically determined. This exercise involved optimization of primer concentration and annealing temperature for each primer. Out of 28 oligonucleotide primers (Bangalore Genei, India) screened, 12 were selected for further analysis based on clarity, scorability, and reproducibility of banding patterns (Table 2). Amplification reactions contained 25 ng template DNA, 200 μM each dNTPs, 2 mM MgCl2, 10 pmol primer, 1 X Taq polymerase buffer and 1 unit of Taq DNA polymerase (Bangalore Genei, India) in a reaction volume of 25 μl. The amplification was performed in programmable gradient Thermal Cycler (BIORAD, USA) with following programme: a pre-denaturation at 94 °C for 3 min followed by 44 cycles of denaturation at 94 °C for 1 min, annealing at gradient temperature for 1 min and extension at 72 °C for 2 min. The final extension was made for 7 min at 72 °C. Amplified products were under electrophoresis in a 1.5 % w/v agarose—1× Tris-borate-EDTA gel at 100 volt. Sizes of the identified bands were determined by comparison with molecular weight-marker low-range DNA ruler (Bangalore Genei, India). Amplicons were scored as discrete variables, using 1 to indicate presence and 0 for absence of band. A binary matrix was obtained by visual scoring of the bands. Amplifications were repeated twice for each primer and only consistent bands were considered for further analyses. Efficiency of discrimination was assessed in terms of the number of polymorphic markers generated and the ability to generate unique band. Pairwise-similarity matrices based on ISSR data were determined using Jaccard’s similarity coefficient [11]. Principal component analysis (PCA) was made by using the binary data of the profile through NTYSIS program.
Results and Discussion
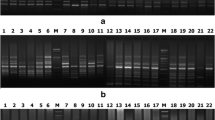
The present investigation reveals that the 48 varieties of rice belonged to two groups. First group having high yielding varieties developed by different Institutes and IR-64 (IR 5657-33-2-1/IR2061-465-1-5-5), an Indica rice variety with distinct desirable agronomic traits. It was included as a check or reference variety. Second group having aromatic rice varieties. In this group, Basmati was included which belongs to an aromatic long grain type of rice (O. sativa) cultivated in different parts of India and Pakistan [12]. Vaughan et al. [13] reported that there are 21 valid species of genus Oryza. Out of them O. sativa (Asian cultivar) and O. glaberrima (African cultivar) are the only cultivated species derived from their perennial wild progenitors O. rufipogon and O. longistaminata respectively. Further, the Asian cultivar has undergone differentiation into three distinct eco-geographical subspecies viz. indica, japonica (temperate japonica) and javanica (tropical japonica). The diversification of O. sativa is not confined with these three sub-species but is differentiated into many more varietal groups largely through human selection under different agroclimatic conditions, cultural practices and quality preferences [14]. Glaszmann [15] reported that the Asian cultivar was differentiated into six varietal groups i.e. Indica, Aus, Ashina, Rayada, Aromatic and Japonica on the basis of isozyme analysis. Basumati rice is a distinct group, well separated from indica and japonica varieties on the basis of morpho-physiological traits as reported earlier [16]. The assessment of genetic diversity and variety diagnostic molecular markers are an essential component in germplasm characterization and conservation. Out of 28 ISSR markers screened, 12 markers were selected to characterize and assess the genetic variability among 48 rice genotypes including four aromatic rice genotypes belonging to indica types because of distinct amplification and highly reproducible bands. The selected ISSR primers yielded 71 bands with an average of 5.92 bands per primer of sizes ranging from 250 bp to 2,000 bp (Fig. 1; Table 2). No unique band was observed, but there were 12 low frequency bands. All other bands (59) were high frequency bands. The ISSR primer (UBC-825) produced the maximum eight bands followed by UBC-872 with seven bands. The lowest number of bands was observed in UBC-864 with four bands. The maximum resolving power (11.417) was observed in primer UBC 825 and the maximum PIC value was obtained by using the ISSR primer (UBC-872). ISSR markers are frequently used for varietal diagnostic purposes in many crop species [17, 18].
The ISSR amplification pattern was used to assess genetic variation among the genotypes by cluster analysis and to detect the varietal diagnostic markers. A dendrogram was plotted using the similarity coefficient derived from ISSR profile data as presented in Fig. 2. The dendrogram was divided into two clusters at 40 % similarity coefficient. The maximum number of genotypes (47) were represented in Cluster I whereas, one genotype ‘CR 1017’, a high yielding variety developed from Central Rice Research Institute formed the cluster II, which was an out-group from the rest of the clusters. Cluster I was again divided into two sub-clusters IA and IB. In sub-cluster IA, there are two rice genotypes whereas 45 genotypes were included in Cluster IB. In sub-cluster IA, these two varieties, “Badami” and “Samant”, with similar pedigree relationship have been grouped together. Both the varieties have medium bold grain with multiple resistance. The sub-cluster IB is further divided into two minor sub-clusters i.e. BI and BII. The minor sub-cluster BI having four varieties of aromatic rice making one group with 60 % similarity coefficient. These four varieties (“Basumati”, “Pumpudibasa”, “Neelabati” and “Kalajerra”) represent aromatic indica rice. The variety “Basumati” and “Pumpudibasa” show 80 % similarity among each other. Both the varieties have 75 % similarity with “Neelabati” and 68 % with “Kalajeera”. They have been developed through natural selection. For these reasons they have been grouped together and placed separately from the other high yielding improved varieties. Further, minor sub-cluster BII is further divided into two sub minor clusters i.e. BIII and BIV. The cluster BIII has 20 varieties and cluster BIV has 21 varieties. In cluster BIII, the variety “Uphar” and “Mandakini” make two individual groups with yield potential 45 q/ha and resistance to blast disease and sheath blight. The variety “Ghanteswari” and “Pathara” make one group with 86 % similarity among themselves. Both the genotypes have same maturity duration of 95 days and equal yield potential of 35 q/ha. The variety “Jogesh” and “Bhanja” have 90 % similarity among themselves with equal yield potential (40 q/ha). These varieties are grown in rain fed and irrigated uplands area with resistant to blast, sheath blast and neck blast disease. These varieties have 86 % similarity with “Ranidhan” and “Bhubana”. The varieties “Parijata” and “MTV 1010” also have 80 % similarity among themselves with some agronomic characteristics like slender grains, photo insensitive, moderately resistant to stem borer and yield potential up to 35 q/ha. The cluster BIV is further divided into two minor clusters, one having 7 varieties and other having 14 varieties. The variety “Lalat” and “IR-64” have 78 % similarity with same yield potential (40 q/ha). Both the varieties have medium slender seeds and multiple resistance to disease and drought. Most of the varieties in this group show 70 % similarity among themselves. The high yielding varieties “Mahalaxmi” (Pankaj/Mahsuri), “Rambha” (Pankaj/W 1263), “Santep Heap” (Pankaj/Sigadis) were placed together in this sub-cluster. All these cultivars have the common parent “Pankaj” and they are the long duration varieties suitable for low land ecosystem. The variety “Jajati” (Rajeswari/T141) and “Urbashi” (Rajeshwari/Jajati) have been grouped together and have originated from the same parent “Rajeshwari”. All these varieties are suitable for cultivation in the eastern part of India. The patterns of diversity revealed by the ISSR primers showed similar correlations between similarity matrices. Most of the ISSR primers especially members of the 5′-anchored dinucleotides could be freely interchanged with no loss in the power to distinguish varietal diversity. These results suggest that the use of different ISSR primers would be able to assess the genetic diversity of rice as reported previously [6, 19].
Principal component analysis indicates that the four aromatic rice varieties (sl. no. 34, 36, 46 and 47 as per Table 1) are making one separate group (Fig. 3). The variety “Badami” and “Samant” are making one out group having the medium bold grain quality with multiple resistant to disease. The differences in ISSR profiles were perhaps due to their adaptability and other significant agronomic characteristics like yield potential, panicle type, seed morphology, photosynthesis efficiency and grain characteristics. The present study showed higher percentage of polymorphism as compared to other molecular markers as reported earlier [20, 21]. The genetic diversity in traditional lowland rice grown in north-east regions using both RAPD and ISSR markers has been reported by Bhuyan et al. [7]. Further, Youssef et al. [9] used both RAPD and ISSR markers to identify the new promising drought tolerant lines of rice. These traits with molecular differences suggest that these rice varieties belong to Indica rice with introgressions from wild rice land races.
Principal component analysis (2-D) of 48 rice varieties using similarity coefficient of ISSR profile data. (Numbers denoting the plotted data points represent the respective rice varieties as listed in Table 1)
In conclusion, ISSR has generated high number of polymorphic markers which can be used in diagnostic fingerprinting studies of rice. Based on the average percentage polymorphism, PIC, Rp, diversity index and marker index, the efficiency of ISSR markers will be helpful for varietal diagnostics and for rice breeding program. In addition, this marker has an important role in securing plant variety rights as the need to protect proprietary germplasm which is likely to increase in the future.
References
Byerlee D (1996). Knowledge-Intensive Crop Management Technologies: Concepts, Impacts, and Prospects in Asian Agriculture. International Rice Research Conference, Bangkok, 3–5 June 1996
Mackill DJ, Zhang Z, Redona ED, Colowit PM (1996) Level of polymorphism and genetic mapping of AFLP markers in rice. Genome 39:969–977
Zhu J, Gale MD, Quarrie S, Jackson MT, Bryan GJ (1998) AFLP markers for the study of rice biodiversity. Theor Appl Genet 96:602–611
McCouch SR, Temnykh S, Lukashova A, Coburn J, Declerck G, Cartinhour S, Harrington S, Thomson M, Septiningsi E, Semon M, Moncada P, Jiming L (2001) Microsatellite markers in rice: abundance, diversity and applications. Rice genetics IV. IRRI, Manila, pp 117–135
Akagi H, Yokozeki Y, Inagaki A, Fujimura T (1997) Highly polymorphic microsatellites of rice consist of AT repeats, and a classification of closely related cultivars with these microsatellite loci. Theor Appl Genet 94:61–67
Joshi SP, Gupta VS, Aggarwal RK, Ranjekar PK, Brar DS (2000) Genetic diversity and phylogenetic relationship as revealed by Inter simple sequence repeat polymorphism in the genus Oryza. Theor Appl Genet 100:1311–1320
Bhuyan N, Borah BK, Sarma RN (2007) Genetic diversity analysis in traditional lowland rice (Oryza sativa L.) of Assam using RAPD and ISSR markers. Curr Sci 93:967–972
Reddy CS, Babu AP, Mallikarjuna BP, Kaladhar K, Sarl N (2009) ISSR markers based on GA and AG repeats reveal genetic relationship among rice varieties tolerant to drought, flood or salinity. J Zhejiang Univ (Sci B) 10:133–141
Youssef MA, Mansour, Solliman SS (2010) Molecular markers for new promising drought tolerant lines of rice under drought stress via. RAPD-PCR and ISSR markers. J Am Sci 6:355–363
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Sneath PHA, Sokal RR (1973) Numerical taxonomy: the principles and practice of numerical classification. W. H. Freeman, San Francisco
Nene YL (1998) Basumati rice: a distinct variety (cultivar) of the Indian subcontinent. Asian Agri-history 2:321–330
Vaughan DA, Morishima H, Kadowaki K (2003) Diversity in the Oryza genus. Curr Opin Plant Mol Biol 6:139–146
Siddiq EA, Vemiredd LR, Nagaraju J (2012) Basmati rices: genetics, breeding and trade. Agric Res 1(1):25–36
Glaszmann JC (1987) Isozymes and classification of Asian rice varieties. Theor Appl Genet 74:21–30
Vairavan S, Siddiq EA, Arunachalam V, Swaminathan MS (1973) A study on the nature of genetic divergence in rice from Assam and North East Himalayas. Theor Appl Genet 43(5):213–221
Raina SN, Rani V, Kojima T, Ogihara Y, Singh KP, Devarumath RM (2001) RAPD and ISSR fingerprints as useful markers for analysis of genetic diversity, varietal identification and phylogenetic relationships in peanut (Arachis hypogaea) cultivars and wild species. Genome 44:763–772
Gorji AM, Poczai P, Polgar Z, Taller J (2011) Efficiency of arbitrarily amplified dominant markers (SCOT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am J Potato Res 88:226–237
Blair MW, Panaud O, McCouch SR (1999) Inter-simple sequence repeat (ISSR) amplification for analysis of microsatellite motif frequency and fingerprinting in rice (Oryza sativa L.). Theor Appl Genet 98:780–792
Galvan MZ, Bornet B, Balatti P, Branchard M (2003) Inter simple sequence repeat (ISSR) markers as a tool for the assessment of both genetic diversity and gene pool origin in common bean (Phaseolus vulgaris L.). Euphytica 132:297–301
Kumar J, Verma V, Qazi GN, Gupta PK (2007) Genetic diversity in Cajanus–Rhynchosia–Flemingia group based on functional markers. Proc Natl Acad Sci India 77(B):369–374
Acknowledgments
The authors wish to acknowledge to Department of Biotechnology, Govt. of India for providing fund for student research under PG HRD program.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kshirsagar, S.S., Samal, K.C., Rabha, M. et al. Identification of Variety Diagnostic Molecular Marker of High Yielding Rice Varieties. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 84, 389–396 (2014). https://doi.org/10.1007/s40011-013-0216-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-013-0216-4