Abstract
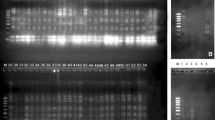
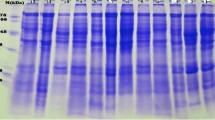
In order to study the genetic diversity among 16 genotypes of Hippophae salicifolia D. Don of Uttarakhand region, random amplified polymorphic DNA (RAPD)–PCR analysis along with protein profiling of seed storage proteins was carried out for selection of superior genotype having high economic importance. A total of 21 RAPD primers were assayed for their specificity in detecting genetic variability in H. salicifolia genotypes, of which 11 RAPD primers were highly reproducible and were found suitable for use in PCR analysis. A total of 198 bands were scored corresponding to an average of 10.8 bands per primer with 138 bands showing polymorphism (73.2 %) with similarity coefficient ranging from 0.011 to 0.999. A dendrogram constructed based on the UPGMA clustering method revealed two major clusters. Cluster-I comprises of 13 cultivars, while cluster-II includes the remaining three cultivars. The cultivar ST-4, ST-5 and ST-16 were quite unique from the remaining cultivars as evident from the dendrogram. The analysis of total seed storage protein (quantitative) and protein profiling (qualitative) among 16 genotypes of H. salicifolia was performed to indentify novel proteins of important functional attributes. SDS-PAGE based generated profiles of seed proteins showed major differences in banding patterns among these genotypes with respect to altitude besides differences in seed protein contents. This study helps in the selection of superior genotype of H. salicifolia having higher economic importance by using developed molecular and protein based markers in Uttarakhand regions.



Similar content being viewed by others
Abbreviations
- RAPD:
-
Random amplified polymorphic DNA
- PCR:
-
Polymerase chain reaction
- SDS:
-
Sodium do-decyl sulphate
- PAGE:
-
Polyacrylamide gel electrophoresis
- SBT:
-
Seabuckthorn
- PIC:
-
Polymorphic information content
- UPGMA:
-
Unweighted pair group method with arithmetic mean
- AFLP:
-
Amplified fragment length polymorphism
- TBSP:
-
Total buffer soluble protein
- SSR:
-
Simple sequence repeats
- SNPs:
-
Single nucleotide polymorphism
- DArT:
-
Diversity arrays technology
References
Gupta SM, Pandey P, Grover A, Ahmed Z (2011) Breaking seed dormancy in Hippophae salicifolia, a high value medicinal plant. Physiol Mol Biol Plants 17(4):403–406
Gupta SM, Ahmed Z (2010) Seabuckthorn (Hippophae salicifolia L.) plant: as source donor of cold tolerant genes for improving high altitude agriculture during cold stress. Res Environ Life Sci 3(3):105–112
Gupta SM, Grover A, Pandey P, Ahmed Z (2012) Female plants of Hippophae salicifolia D. Don are more responsive to cold stress than male plants. Physiol Mol Biol Plant 18(4):377–380
Gupta SM, Gupta AK, Ahmed Z, Kumar A (2011) Antibacterial and antifungal activity in leaf, seed extract and seed oil of seabuckthorn (Hippophae salicifolia D. Don) plant. J Plant Pathol Microbiol 2:105. doi:10.4172/2157-7471.1000105
Yadav VK, Sah VK, Singh AK, Sharma SK (2006) Variations in morphological and biochemical characters of seabuckthorn (Hippophae salicifolia D. Don) populations growing in Harsil area of Garhwal Himalaya in India. Trop Agric Res Ext 9:1–7
Persson H (2001) Estimating genetic variability in horticultural crop species at different stages of domestication. Doctoral thesis, ISSN 1401-6249, ISBN 91-576-5838-2
Farooq S, Azam F (2002) Plant breeding-II. Some pre-requisites for use. Pak J Biol Sci 5(10):1141–1147
Asad HS, Syed DA, Ishtiaque K, Farhat B, Lutful H, Stephen RP (2009) Evaluation of phylogenetic relationship among seabuckthorn (Hippophae rhamnoides L. spp. Turkestanica) wild ecotypes from Pakistan using amplified fragment length polymorphism (AFLP). Pak J Bot 41(5):2419–2426
Sezai E, Emine O, Nalan Y, Güleray A (2008) Comparison of seabuckthorn genotypes (Hippophae rhamnoides L.) based on RAPD and FAME data. Turk J Agric For 32:363–368
Bartish N IV, Jeppsson, Bartish GI (2000) Inter- and intraspecific genetic variation in Hippophae (Elaeagnaceae) investigated by RAPD markers. Plant Syst Evol 225(1–4):85–101
Ahmad SD, Kamal M (2002) Morpho-molecular characterization of local genotypes of Hyppophae rhamnoides L. spp. Turkestanica Int J Agric Biol 5:10–13
Xin Z, Chen J (2012) A high throughput DNA extraction method with high yield and quality. Plant Methods 8:26
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4326
Rohlf FJ (1997) NTSYS-pc numerical taxonomy and multivariate analysis system, version 2.0. Exeter, New York
Yap IV, Nelson RJ (1996) Winboot: a program for performing bootstrap analysis of binary data to determine the confidence of UPGMA-based dendrograms. IRRI, Manilla
Tatham AS, Fido RJ, Moore CM, Quesada DD, Kuzmicky DD, Keen JN, Shewry PR (1996) Characterisation of the major prolamins of Tef (Eragrostis tef) and Finger millet (Eleusine coracana). J Cereal Sci 24:65–71
Singh D, Sirohi A, Sharma VS, Kumar V, Gaur A, Dhaudhary R, Choudhary N (2006) RAPD based identification of farmers collections of Kalanamak and traditional varieties of Basmati rice. Indian J Crop Sci 1:102–105
Brussell JD, Woycott M, Chappill JA (2005) Arbitrarily amplified DNA markers as characters for phylogenetic inference. Prospect Plant Eco Evol Syst 7:3–26
Ahmad SD, Jasra AW, Imtiaz A (2003) Genetic diversity in Pakistani genotypes of Hippophae rhamnoides L. spp. Turkestanica Int J Agric Biol 5:10–13
Bradford MM (1976) Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Acknowledgments
The authors acknowledge the Dean, Hill campus, G.B. Pant Agriculture and Technology Ranichauri for providing seed materials. They also thank to Dr. V.K. Yadav, Hill campus Ranichauri, G.B. Pant Agriculture and Technology for providing the passport in formation of H. salicifolia genotype used into the present work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sharma, N., Gupta, S., Singh, S. et al. Use of Molecular and Protein Based Markers for Accessing Genetic Diversity Among Hippophae salicifolia D. Don Genotypes of Uttarakhand. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 84, 47–54 (2014). https://doi.org/10.1007/s40011-013-0196-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-013-0196-4