Abstract
Background
Influenza is an important public health problem. The aim of this study was to evaluate and compare the sensitivity and specificity of three rapid diagnostic tests (SEKISUI, QuickVue Influenza A + B, and SD BIOLINE) for novel swine-origin influenza viruses (S-OIV) and seasonal influenza.
Materials and methods
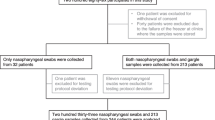
A total of 210 nasopharyngeal swabs from unique clinical specimens were previously tested by real-time reverse transcription polymerase chain reaction (RT-PCR) assay and tested again in this study.
Results and discussion
Of these, 164 (78%) were influenza A-positive and 46 (22%) were influenza A-negative by RT-PCR. From 115 positive swabs, 80 (69.6%), 66 (57.4%), and 46 (40.0%) showed S-OIV by SEKISUI, QuickVue Influenza A + B, and SD BIOLINE, respectively. Specific positive and negative predictive values of these three commercial rapid tests were all 100%. Therefore, positive rapid influenza virus diagnosis does not require an RT-PCR confirmatory test. Conversely, only negative rapid influenza virus diagnosis should be evaluated. The SEKISUI test would be a useful diagnostic tool for screening clinical samples for influenza. Concerning the various specimen types, among 25 patients with RT-PCR-proven S-OIV infection, influenza was identified in sputum (21/25; 84.0%) and nasopharyngeal swab (15/25; 60.0%) specimens, but in only 36.0% (9/25) of throat swab specimens. Sputum and nasopharyngeal swab specimens were the most predictive of influenza virus infection, while throat swab specimens were the least predictive of influenza virus infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Novel H1N1 swine-origin influenza A virus (S-OIV) is a contagious respiratory viral illness, which is transmitted easily from human to human and leads to global spread [1, 5–7, 21, 33, 34]. As a consequence, this virus has emerged in resource-limited countries [5, 14, 22, 32, 34]. While a variety of different diagnostic approaches have been projected, the Centers for Disease Control and Prevention (CDC) currently recommends that suspected clinical S-OIV infection be confirmed by real-time reverse transcription polymerase chain reaction (RT-PCR) or viral culture. However, a recent number of studies have demonstrated the sensitivity and specificity of RT-PCR for the detection of influenza viruses, and influenza experts have recommended that RT-PCR be regarded as the new diagnostic standard [3, 17, 18, 20, 23–25, 29].
The exact characterization of influenza A viruses is fundamental for diagnostics. Nevertheless, the large pandemic of this S-OIV has focused the need to develop and introduce rapid influenza diagnostic tests (RIDTs) [5, 6, 10, 12, 31, 32]. These tests must be able to rapidly and efficiently detect these viruses in respiratory clinical specimens. The clinical samples can be taken from deep within the nostrils (nasal swab), nasopharynx (nasopharyngeal swab), nasopharyngeal aspirate, or throat [4, 6–10, 12, 14, 16, 18, 25, 27, 32]. Previous reports have demonstrated that RIDTs with sensitivities of 39–80% show high specificities of approximately 95–100%, depending upon the trade mark and the level of influenza virus in the collected sample [4, 6–12, 14–17, 19, 24–29]. RIDTs permit a point-of care diagnosis to be completed within 30 min in patients with influenza-like illness [9, 10, 12].
The rapid detection of a certain virus is essential for proper treatment, disease containment, and the timely implementation of infection control in a resource-limited setting [5, 13, 14, 26, 32–34]. Due to the high costs and complicated procedures associated with RT-PCR, the clinical sensitivity, specificity, positive predictive values (PPVs) and negative predictive values (NPVs) of the commercial RIDTs, SEKISUI, QuickVue Influenza A + B, and SD BIOLINE, were evaluated for the detection of influenza viruses in a primary healthcare setting in Thailand. And, finally, when evaluating the relative performance of RIDTs, it is essential to consider the type of clinical specimens; thus, differential detection of influenza viruses in different respiratory specimens was also performed in this study.
Materials and methods
Clinical specimens for comparative study
A total of 210 clinical specimens, which had been previously tested by RT-PCR assay (RT-PCR 480 Roche Diagnostic, Thailand), were used in this study. The age of the patients ranged from 6 months to 15 years, most were children and young adults, and the median age was 5 years. All nasopharyngeal specimens were obtained from patients who had CDC-defined influenza-like symptoms, and had visited Ramathibodi Hospital, Mahidol University, during the outbreak of the 2009 pandemic influenza A (H1N1) virus infection in Thailand. These nasopharyngeal specimens were used freshly for all consequent procedures and were not frozen prior to usage.
In a clinical study comparing the impact of specimen types on the rapid diagnostic tests, a total of 75 clinical specimens were collected simultaneously from 25 patients experiencing influenza-like symptoms with RT-PCR-proven S-OIV infection. Clinical specimens from sputum (n = 25), nasopharyngeal swab (n = 25), and throat swab (n = 25) specimens were detected by the rapid diagnostic test SEKISUI (Genzyme, USA), as described in the package insert. This study was approved by the Institutional Review Board (IRB) at Ramathibodi Hospital.
Viral RNA extraction
All samples were placed in viral transport media (1 mL) and immediately submitted to the Virology and Molecular Microbiology Unit, Ramathibodi Hospital, and the total RNA was then automatically extracted using the NucliSENS® easyMAG™ apparatus (bioMérieux, the Netherlands), according to the manufacturer’s instructions. Briefly, 200 μl of nasopharyngeal solution was added to 2 mL of lysis buffer and incubated for 10 min at room temperature, and 50 μl of nucleic acid was eluted after automatic magnetic separation.
Comparative analysis of rapid diagnostic tests
To evaluate the analytical sensitivity and specificity for three rapid commercial kits in patients with pandemic (H1N1) 2009 virus, 210 RT-PCR-confirmed clinical specimens were used. The three rapid commercial kits for influenza A viruses were [1] SD BIOLINE influenza antigen (Standard Diagnostics Inc., Korea), [2] QuickVue Influenza A + B (Quidel Corporation, San Diego, CA), and [3] SEKISUI (Genzyme, USA). All tests were carried out according to the manufacturers’ instructions and the laboratory-validated protocols. Briefly, 100 μl of nasopharyngeal supernatant specimens were added to lysis reagent and then placed on a dip strip in the prepared sample, with the arrow on the stick pointing downward. The test results were interpreted after the stick was dipped in solution for exactly 10 min. A positive conclusion was considered if, at 10 min, a pink line had appeared to indicate a positive influenza A and B result.
Quantitative RT-PCR, standard curve, and viral titer in clinical specimens
Among 164 clinical specimens, which had been previously tested and confirmed positive by real-time RT-PCR, the virus concentration was determined without thawing, using real-time RT-PCR (LightCycler Real-Time PCR 2.0, Roche Diagnostic, Thailand) for the Matrix (M) gene. Each positive RT-PCR clinical specimen was characterized by the Ct value demonstrated in the universal influenza A. Subsequently, ten samples with Ct values ranging from 21 to 22 were selected. The relative sensitivity of the three rapid tests was compared by analyzing twofold serial dilutions (1:2, 1:4. 1:8, 1:16, 1:32, and 1:64) of selected samples. The sensitivity of rapid tests was determined as the percentage of positive specimens among the number of serial dilutions.
For the copy number determination, a positive control template was amplified by real-time RT-PCR. The standard curve was created according to the reaction, in which tenfold serial dilutions ranged from 101 to 106 copies of referent standard influenza A virus. Therefore, the virus concentration could be extrapolated based on the standard curve.
Statistical analysis
The sensitivity, specificity, PPV, and NPV were calculated using standard formulas. Agreement between RT-PCR and the rapid tests was calculated using the kappa statistic. The Pearson Chi-square test was used to compare the results in this study. Statistical analyses were performed using the Minitab program. A P value of <0.05 was considered to be statistically significant.
Results
Comparative analysis of the rapid diagnostic tests for the detection of influenza A virus
A total of 210 nasopharyngeal swabs, which had been previously tested by real-time RT-PCR, were retested in this study. Among them, the number of positive cases was 164 (78%) samples, while 46 (22%) were negative for influenza A. Of all 164 RT-PCR-confirmed positive cases, 114, 96, and 74 samples could be detected by using SEKISUI, QuickVue, and SD BIOLINE, respectively (Table 1). Conversely, in 46 negative RT-PCR tests, no positive samples were found in any of the rapid tests. The specificity, PPV, and NPV were 100% for all three rapid tests. The sensitivity of all three rapid assays in comparison with real-time RT-PCR was 69.51% (SEKISUI), 58.54% (QuickVue), and 45.12% (SD BIOLINE). The sensitivity, specificity, PPV, and NPV of the rapid tests are presented in Table 2.
Comparative analysis of rapid diagnostic tests for S-OIV and seasonal influenza virus
Of these 164 influenza A-positive specimens, 115 (70%) were positive for S-OIV, while 49 (30%) were positive for seasonal influenza A (H1N1 and H3N2) virus by the real-time RT-PCR test. The ability of the three commercial rapid influenza kits for the detection of S-OIV were as follows: 46/115 (40.0%), 66/115 (57.4%), and 80/115 (69.6%) for SD BIOLINE, QuickVue, and SEKISUI, respectively. Therefore, SEKISUI had a more significant sensitivity (P < 0.05) than SD BIOLINE and QuickVue. For the 49 seasonal influenza samples, 57.1% (n = 28), 61.2% (n = 30), and 69.4% (n = 34) were detected by SD BIOLINE, QuickVue, and SEKISUI, respectively. Overall, the sensitivity of the rapid tests was not significantly different (P > 0.05) for the detection of seasonal influenza samples, except for SD BIOLINE (P < 0.05). Table 3 presents a comparative analysis of the rapid diagnostic tests for S-OIV and seasonal influenza.
Rapid detection of influenza A in clinical specimens and viral titers
Among the 164 PCR-confirmed positive cases, ten samples of influenza A, with high viral titers, were used to determine the sensitivity of the three commercial rapid tests. The selected real-time RT-PCR-confirmed positive clinical specimens, with Ct 21–22 (median concentration of 82,150 RNA copies/mL), were titrated against the Matrix gene level of virus stocks. For twofold serial dilutions of clinical samples (nasal swabs), starting at undilution, 1:2, 1:4, 1:8, 1:16, 1:32, and 1:64 were prepared. The concentration of virus in the samples was determined from RNA extract without thawing the original samples, using real-time RT-PCR. From a series of twofold dilutions, all virus stocks yielded an endpoint (1:64) at approximately 1,250 copies/mL.
The sensitivity of each of the three rapid tests, with the twofold dilution of clinical specimens, is detailed in Table 4. Using the SEKISUI rapid test for the detection of influenza A, 10 (100%) samples gave positive results until 1:4, whereas QuickVue and SD BIOLINE gave positive results until 1:2 and undilution, respectively. Of ten RT-PCR-confirmed positive samples, the number of positive results per series of twofold dilution by SEKISUI and QuickVue for the detection of all influenza A strains were as follows: undilution (10/10; 100%), 1:2 (10/10; 100%), 1:4 (10/10; 100% and 9/10; 90%), 1:8 (7/10; 70%), 1:16 (5/10; 50% and 4/10; 40%), and 1:32 (2/10; 20%). Nonetheless, the ability of SD BIOLINE to detect a series of twofold dilutions was different, as follows: undilution (10/10; 100%), 1:2 (9/10; 90%), 1:4 (6/10; 60%), 1:8 (4/40; 40%), and 1:16 (2/10; 20%).
Examining the impact of specimen type on rapid diagnostic tests
The percentages of positive specimens detected by rapid tests were calculated for each specimen type. Sputum specimens most accurately predicted influenza virus infection, with 84.0% (21/25) of infected patients being identified. Nasopharyngeal swab specimens were the second best sample, with 60.0% (15/25) of those infected being identified, whereas throat swab specimens 9/25; 36.0%) were the least predictive of influenza virus infection and were significantly less predictive than sputum (P < 0.01) and nasopharyngeal swab (P < 0.05) specimens to detect influenza virus or viral antigen from patients considered to be positive for S-OIV.
Discussion
Rapid and accurate detection is fundamental for diagnosing influenza A viruses in order to provide evidence for quarantine and medication in controlling further outbreaks [5, 10, 13, 26, 33]. This study was the first designed to evaluate and compare the performances of three rapid diagnostic tests (SEKISUI, QuickVue Influenza A + B, and SD BIOLINE) for the early detection of the novel swine-origin influenza viruses (S-OIV) and seasonal influenza (H1N1 and H3N1) in clinical specimens during the pandemic outbreak of novel H1N1 in developing countries [6–9, 12, 14, 15, 27, 28, 31, 32, 35].
The overall sensitivity of SEKISUI, QuickVue Influenza A + B, and SD BIOLINE, when compared to RT-PCR, was 69.5, 58.5, and 45.1%, respectively. The sensitivity of the SD BIOLINE test was significantly lower than that in the SEKISUI assay for the detection of influenza A (P < 0.05, by Fisher’s exact test). In agreement with our report, Gordon et al. [14] described the clinical sensitivity of the rapid test for the detection of influenza viruses in a primary healthcare setting. The sensitivity of QuickVue for the detection of children with fever was 68.5%. Nonetheless, a former study observed only 27% clinical sensitivity for seasonal influenza detection for the same assay [30]. Moreover, Drexler et al. [11] showed a sensitivity of merely 11% for the detection of influenza from clinical specimens when using a rapid test.
The lower sensitivity of assays with clinical specimens from the stored group compared to those in the fresh group was probably due to virus degradation from stored clinical specimens [13]. In our study, no specimens from storage were used to test the rapid assays. This may have contributed to the higher sensitivity of detection compared to previous studies. Rapid diagnostic tests can help in the diagnosis and management of patients who present with signs and symptoms compatible with influenza [2, 7, 14, 28, 32]. However, the reliability of rapid tests was considered in the importance of specimen type, quality, sensitivity, and specificity of the kit. Clinicians may consider using rapid diagnostic tests as part of their evaluation of patients with signs and symptoms compatible with influenza, but the results should be interpreted with caution.
However, specificity for the detection of all influenza A subtypes was 100% and no false-positive results were found for all rapid assays. Therefore, positive rapid test results performed well in predicting confirmed infection during the outbreak of influenza viruses. Even so, the NPVs were 47.9, 40.4, and 33.8% for SEKISUI, QuickVue, and SD BIOLINE, respectively. Importantly, the results from this study established that the rapid test could not be used to rule out influenza A (S-OIV and seasonal viruses) infection in individuals with a negative result. Conversely, the positive results of a rapid test (PPV = 100%) could be used to screen influenza A infection in a resource-limited setting during an outbreak.
Moreover, all specimens were obtained from patients meeting the CDC definition of influenza-like illness [4], so it is more reasonable to compare the performance characteristics for various rapid tests in the detection of seasonal influenza strains and S-OIV. The SEKISUI assay showed higher sensitivity for the detection of both S-OIV (80/115; 69.6%) and seasonal influenza (34/49; 69.4%) when compared to QuickVue (57.4% for S-OIV and 61.2% for seasonal influenza) and SD BIOLINE (40.0% for S-OIV and 57.1% for seasonal influenza). The findings in this study are comparable to previously reported observations that rapid tests had average sensitivity in detecting influenza A in clinical specimens [8, 9, 14–16, 26–28, 32, 35].
Recently, several studies have reported varying sensitivities of rapid tests from the viral titer of specimens [4, 9, 14, 15]. Therefore, the relative sensitivity was observed to demonstrate the effect of virus concentration on the detection sensitivity of rapid commercial kits by analyzing twofold serial dilutions (1:2, 1:4. 1:8, 1:16, 1:32, and 1:64) of selected samples using the three rapid tests. The virus concentration was estimated from cycle threshold numbers (CT) of undiluted specimens. The decreased sensitivity of the three rapid tests was consistent with the twofold series dilution of virus stock. The sensitivity (≥50% of ten samples) for each test, SEKISUI, QuickVue, and SD BIOLINE, reached approximately 5,000, 10,000, and 20,000 copies/mL, respectively. Our result proposes that the diversity of sensitivity was positively associated with the titer of viruses in the clinical samples.
Although different clinical specimens are considered as proper for rapid detection, not every clinical specimen type yields comparable outcome. Practically, when evaluating the performance of different rapid diagnostic tests for influenza, it is essential to consider the type of clinical specimens. In this study, 75 clinical specimens with RT-PCR-proven S-OIV infection were identified in sputum (84.0%) and nasopharyngeal swab (60.0%) specimens, but in only 36.0% of throat swab specimens. As a result, sputum and nasopharyngeal swab specimens should be obtained to diagnostic accuracy, whereas throat swabs are the least sensitive and specific. This possibly reveals the higher virus loads found in sputum and nasopharyngeal swab specimens compared to throat swabs.
Importantly, improper specimen collection and the types of clinical specimen tested are partial factors that might decrease the performance of rapid tests [2, 4, 14, 28, 30]. Moreover, inappropriate specimen handling is an important reason for causing a decrease in the sensitivity of assays [4]. To evaluate assay performance on clinical specimens, we used an experienced physician to collect nasopharyngeal swabs from patients with CDC-defined influenza-like illness. This physician was trained in the procedure at Ramathibodi Hospital, a center with the highest number of reported cases during the S-OIV outbreak in Thailand. In addition, other factors that could affect the sensitivity of rapid assays are the quality of specimens before testing and the interval from symptom onset to specimen collection [4]. However, this study was performed in a clinical setting, where individuals were usually present on the day of symptom onset.
In conclusion, the rapid screening tests used in this study were only useful in samples that tested positive for influenza A viruses, although negative samples could not be ruled out regarding infection. However, the moderate sensitivity and NPV of these tests emphasize the limitation of using a rapid test alone to establish diagnosis and clinical decision-making. Even though rapid tests can be performed in a primary healthcare setting, and have been shown as the key to outbreak control, improvement to the sensitivity of rapid testing is needed.
References
Beck ET, Jurgens LA, Kehl SC, Bose ME, Patitucci T, LaGue E, Darga P, Wilkinson K, Witt LM, Fan J, He J, Kumar S, Henrickson KJ. Development of a rapid automated influenza A, influenza B, and respiratory syncytial virus A/B multiplex real-time RT-PCR assay and its use during the 2009 H1N1 swine-origin influenza virus epidemic in Milwaukee, Wisconsin. J Mol Diagn. 2010;12:74–81.
Belizario VY, Pasay CJ, Bersabe MJ, de Leon WU, Guerrero DM, Bugaoisan VM. Field evaluation of malaria rapid diagnostic tests for the diagnosis of P. falciparum and non-P. falciparum infections. Southeast Asian J Trop Med Public Health. 2005;36:552–61.
Centers for Disease Control and Prevention (CDC). 2009. CDC protocol of realtime RTPCR for influenza A(H1N1). Available online at: http://www.who.int/csr/resources/publications/swineflu/realtimeptpcr/en/index.html.
Centers for Disease Control and Prevention (CDC). Performance of rapid influenza diagnostic tests during two school outbreaks of 2009 pandemic influenza A (H1N1) virus infection—Connecticut, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:1029–32.
Centers for Disease Control and Prevention (CDC). Outbreak of swine-origin influenza A (H1N1) virus infection—Mexico, March–April 2009. MMWR Morb Mortal Wkly Rep. 2009;58:467–70.
Cunha BA, Syed U, Mickail N, Strollo S. Rapid clinical diagnosis in fatal swine influenza (H1N1) pneumonia in an adult with negative rapid influenza diagnostic tests (RIDTs): diagnostic swine influenza triad. Heart Lung. 2010;39:78–86.
Cunha BA, Syed U, Stroll S, Mickail N, Laguerre M. Winthrop-University Hospital Infectious Disease Division’s swine influenza (H1N1) pneumonia diagnostic weighted point score system for hospitalized adults with influenza-like illnesses (ILIs) and negative rapid influenza diagnostic tests (RIDTs). Heart Lung. 2009;38:534–538.
Dale SE, Mayer C, Mayer MC, Menegus MA. Analytical and clinical sensitivity of the 3M rapid detection influenza A+B assay. J Clin Microbiol. 2008;46:3804–7.
de la Tabla VO, Antequera P, Masiá M, Ros P, Martin C, Gazquez G, Buñuel F, Sánchez V, Robledano C, Gutiérrez F. Clinical evaluation of rapid point-of-care testing for detection of novel influenza A (H1N1) virus in a population-based study in Spain. Clin Microbiol Infect (in press).
de La Rocque F, Lécuyer A, Wollner C, d’Athis P, Pecking M, Thollot F, Cohen R. Impact of influenza rapid diagnostic tests (IRDT) on the diagnosis of influenza and on the management of influenza in children in ambulatory pediatric setting. Arch Pediatr. 2009;16:288–93.
Drexler JF, Helmer A, Kirberg H, Reber U, Panning M, Müller M, Höfling K, Matz B, Drosten C, Eis-Hübinger AM. Poor clinical sensitivity of rapid antigen test for influenza A pandemic (H1N1) 2009 virus. Emerg Infect Dis. 2009;15:1662–4.
Fernandez C, Cataletto M, Lee P, Feuerman M, Krilov L. Rapid influenza A testing for novel H1N1: point-of-care performance. Postgrad Med. 2010;122:28–33.
Frisbie B, Tang YW, Griffin M, Poehling K, Wright PF, Holland K, Edwards KM. Surveillance of childhood influenza virus infection: what is the best diagnostic method to use for archival samples? J Clin Microbiol. 2004;42:1181–4.
Gordon A, Videa E, Saborio S, López R, Kuan G, Reingold A, Balmaseda A, Harris E. Performance of an influenza rapid test in children in a primary healthcare setting in Nicaragua. PLoS One. 2009;4:e7907.
Herzum I, Lutz T, Koch F, Geisel R, Gehrt A. Diagnostic performance of rapid influenza antigen assays in patients infected with the new influenza A (H1N1) virus. Clin Chem Lab Med. 2010;48:53–6.
Hurt AC, Alexander R, Hibbert J, Deed N, Barr IG. Performance of six influenza rapid tests in detecting human influenza in clinical specimens. J Clin Virol. 2007;39:132–5.
Karre T, Maguire HF, Butcher D, Graepler A, Weed D, Wilson ML. Comparison of Becton Dickinson Directigen EZ Flu A+B test against the CDC real-time PCR assay for detection of 2009 pandemic influenza A/H1N1 virus. J Clin Microbiol. 2010;48:343–4.
Koenig M, Kosha S, Hickman M, Heath D, Riddell S, Aldous W. Detection of influenza virus from throat and pharyngeal swabs with a nested duplex light cycler RT-PCR. Diag Microbiol Infect Dis. 2003;46:35–7.
Kok J, Blyth CC, Foo H, Patterson J, Taylor J, McPhie K, Ratnamohan VM, Iredell JR, Dwyer DE. Comparison of a rapid antigen test with nucleic acid testing during cocirculation of pandemic influenza A/H1N1 2009 and seasonal influenza A/H3N2. J Clin Microbiol. 2010;48:290–1.
Liolios L, Jenney A, Spelman D, Kotsimbos T, Catton M, Wesselingh S. Comparison of a multiplex reverse transcription-PCR-enzyme hybridization assay with conventional viral culture and immunofluorescence techniques for the detection of seven viral respiratory pathogens. J Clin Microbiol. 2001;39:2779–83.
Michaelis M, Doerr HW, Cinatl J Jr. An influenza A H1N1 virus revival—pandemic H1N1/09 virus. Infection. 2009;37:381–9.
Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team, Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15.
Pabbaraju K, Wong S, Wong AA, Appleyard GD, Chui L, Pang X-L, Yanow SK, Fonseca K, Lee BE, Fox JD, Preiksaitis JK. Design and validation of real-time reverse transcription-PCR assays for detection of pandemic (H1N1) 2009 virus. J Clin Microbiol. 2009;47:3454–60.
Rockett RJ, Bialasiewicz S, Whiley DM, Bletchly C, Faux CE, Lambert SB, Nimmo GR, Nissen MD, Sloots TP. A novel duplex real-time reverse transcription PCR assay for the detection of influenza A and the novel influenza A (H1N1) strain. Viruses. 2009;1:1204–8.
Sánchez-Torrent L, Triviño-Rodriguez M, Suero-Toledano P, Claret-Teruel G, Muñoz-Almagro C, Martínez-Sánchez L, Jordán-García I, Garcia-Garcia JJ. Novel influenza A (H1N1) encephalitis in a 3-month-old infant. Infection. 2010;38:227–9.
Sambol AR, Abdalhamid B, Lyden ER, Aden TA, Noel RK, Hinrichs SH. Use of rapid influenza diagnostic tests under field conditions as a screening tool during an outbreak of the 2009 novel influenza virus: practical considerations. J Clin Virol. 2010;47:229–33.
Scansen KA, Bonsu BK, Stoner E, Mack K, Salamon D, Leber A, Marcon MJ. Comparison of polyurethane foam to nylon flocked swabs for collection of secretions from the anterior nares in performance of a rapid influenza virus antigen test in a pediatric emergency department. J Clin Microbiol. 2010;48:852–6.
Suntarattiwong P, Jarman RG, Levy J, Baggett HC, Gibbons RV, Chotpitayasunondh T, Simmerman JM. Clinical performance of a rapid influenza test and comparison of nasal versus throat swabs to detect 2009 pandemic influenza A (H1N1) infection in Thai children. Pediatr Infect Dis J. 2010;29:366–7.
Terletskaia-Ladwig E, Eggers M, Meier S, Leinmüller M, Schneider F, Schmid M, Enders M. Laboratory-based assessment of influenza in German ambulant patients from 1998 to 2008. Infection. 2009;37:401–6.
Uyeki TM, Prasad R, Vukotich C, Stebbins S, Rinaldo CR, Ferng YH, Morse SS, Larson EL, Aiello AE, Davis B, Monto AS. Low sensitivity of rapid diagnostic test for influenza. Clin Infect Dis. 2009;48:e89–92.
Vasoo S, Stevens J, Singh K. Rapid antigen tests for diagnosis of pandemic (Swine) influenza A/H1N1. Clin Infect Dis. 2009;49:1090–3.
Watcharananan S, Kiertiburanakul S, Chantratita W. Rapid influenza diagnostic test during the outbreak of the novel influenza A/H1N1 2009 in Thailand: an experience with better test performance in resource limited setting. J Infect. 2010;60:86–7.
Woo PCY, Chiu SS, Seto WH, Peiris M. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J Clin Microbiol. 1997;35:1579–81.
World Health Organization (WHO). 2009. Global Alert and Response (GAR): Pandemic (H1N1) 2009. Available online at: http://www.who.int/csr/disease/swineflu/en/.
Yoo Y, Sohn JW, Park DW, Kim JY, Shin HK, Lee Y, Choung JT, Lee CK, Kim MJ. Clinical evaluation of the SD Bioline influenza virus antigen test for rapid detection of influenza viruses A and B in children and adults during the influenza season. Clin Vaccine Immunol. 2007;14:1050–2.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
C. Sukasem and V. Pongthanapisith are the co-first authors of this paper.
Rights and permissions
About this article
Cite this article
Pongthanapisith, V., Sukasem, C., Premchaiporn, K. et al. Clinical performance of three rapid diagnostic tests for influenza virus in nasopharyngeal specimens to detect novel swine-origin influenza viruses. Infection 39, 105–111 (2011). https://doi.org/10.1007/s15010-011-0092-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-011-0092-x