Abstract
Novel Schiff base ligand was prepared by the condensation of 2,2-(ethylenedioxy)bis(ethylamine) and imidazole-2-carboxaldehyde in a 1:2 ratio, and its complexes with Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), and Cd(II) metal ions were synthesized in a 1:1 ratio. Various methods were used to identify the Schiff base ligand and complexes. Characterization techniques such as infrared, UV, and 1H NMR spectral tests, elemental analysis, molar conductivity, magnetic properties, thermal analysis, BET surface area, and theoretically by DFT were used to help further understanding of the complex structures. For all complexes, the ligand behaved as a neutral tetradentate ligand with NOON donor atoms and spectroscopic studies indicated an octahedral structure. The electrolytic behavior of the complexes was revealed by the molar conductivity measurements. In addition, thermogravimetric analysis was used to investigate the decomposition of the complexes and Schiff base ligand. According to the results obtained, the antibacterial activity of the metal complexes is higher than that of the Schiff base ligand against one or more bacterial species. The results showed that Cd(II) complex exhibits enhanced activity against Aspergillus flavus and Ni(II) complex against Candida albicans than the standard drug ketoconazole. The Mn(II) complex had stronger activity against human breast cancer cell line (MCF-7) than the other complexes. Antiviral drug development is critical in the latest outbreak of a novel coronavirus (SARS-CoV-2). The new coronavirus was screened using molecular docking.
Similar content being viewed by others
Introduction
The Schiff bases derived from imidazole are easily produced by combining the corresponding carboxaldehyde imidazole with an amine derivative during a condensation. Generally, aromatic aldehydes and aromatic amines generate higher yields than aliphatic precursors for Schiff bases [1]. The most promising advancement in the field of medicinal chemistry is due to the introduction of heterocyclic compounds which performed an important function in regulating the biological activities [2, 3].
The electron dense aryl groups are responsible for stabilizing the imine bond during electron delocalization. Because of its ease of synthesis and the widespread presence of imidazole moieties in enzymes, proteins, and pharmaceuticals, it is a popular option in biological research. Schiff base ligands also can coordinate and stabilize metals in a broad variety of oxidation states. The transition metal complexes having oxygen and nitrogen donor Schiff bases keep unusual configuration, structural liability, and are potentially sensitive to the molecular environment. Transition metal Schiff base complexes are widely investigated within the scientific literature [4, 5].
With these details in mind, it had been decided to synthesize metal complexes of Schiff base ligands to reinforce their biological properties. The hexacoordinated Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), and Cd(II) complexes were synthesized using the NOON tetradentate Schiff base ligand, which was generated by combining 2,2-(ethylenedioxy)bis(ethylamine) and imidazole-2-carboxaldehyde. These metal complexes exhibited remarkable antimicrobial and anticancer properties, suggesting that they could be useful in medicinal chemistry [6]. In recent years, a considerable number of tetradentate salen-type Schiff base compounds, especially those have a NOON donor sets, can be used as ligands to obtain metal complexes that can serve as successful models of biological compounds [7, 8]. Schiff bases with oxygen and nitrogen donor atoms operate as good chelating agents for both transition and non-transition metals [9].
Schiff base ligands are derived from heterocyclic compounds with two distinct forms of functional elements in the ring, such as amino and carbonyl groups. Heterocyclic compounds are synthesized with potential medicinal properties and medical significance; as a result, these compounds have a wide range of industrial applications in biological products [10].
Schiff base ligands were a class of compounds that were commonly used as enzyme inhibitors and had a wide range of pharmacological uses. These compounds are used to be distinct in the field of microbiological research for tumor chemotherapy and will mask the antitumor activity against certain Gram-negative bacteria. Anticancer properties of metal complexes made against MCF7 cell line [10].
The novel coronavirus (2019-nCoV) has recently been discovered in Hubei Province, P.R. China. On January 10, 2020, the whole-genome sequence of 2019-nCoV was published for the first time. 2019-nCoV infects a wide variety of mammals, as well as humans. The probability of transmission from animals to humans is determined by this feature of transmission. The spread of 2019-nCoV has sparked widespread concern and concern humans have experienced two coronavirus-related crises since 2003. The Middle East Respiratory Syndrome Coronavirus (MERS-CoV) appeared in the Arabian Peninsula in 2012, with a fatality rate of 35%, after the severe acute respiratory syndrome coronavirus (SARS-CoV) broke out in 2003 [11].
The aim of this research was to create a new Schiff base ligand and its metal complexes. The structures of the prepared compounds were identified using a variety of characterization methods. Antibacterial, antifungal, and anti-breast cancer activities were also tested in vitro. The likely binding of the prepared compounds with the crystal structure of the unliganded active site of the SARS-CoV-2 main protease (2019-nCoV, coronavirus disease 2019, COVID-19) (PDB ID: 6Y84) was demonstrated [10].
Experimental
All the chemicals used were analytical reagent grade (AR) and of the highest purity. The chemicals used were imidazole-2-carboxaldehyde (Sigma-Aldrich), 2,2-(ethylenedioxy)bis(ethylamine) (Sigma-Aldrich), CrCl3·6H2O, MnCl2·4H2O, and FeCl3·6H2O (Sigma-Aldrich), CoCl2·6H2O, NiCl2·6H2O, CuCl2·2H2O, and CdCl2 (Merck). Ethyl alcohol (95%) and N,N-dimethylformamide were used as organic solvents (DMF). In all the preparations, double-distilled water was widely used.
Solutions
Stock solutions (1 × 10−3 M) and dilute solutions (of 1 × 10−4 M and 1 × 10−5 M) of the Schiff base ligand (L) and its metal complexes were prepared in DMF solvent, and they were used for conductivity and UV–Vis spectra measurements.
Solution of anticancer study
In the appropriate volume of DMF, a fresh stock solution (1 × 10−3 M) of Schiff base ligand (0.12 × 10−2 g L−1) was prepared. Cryopreservation of cells was done with DMSO. The medium used was RPMI-1640. The human tumor cell line was cultured and kept in this medium. The medium was provided in the form of a powder. It was prepared in the following manner:
10.4 g of medium was weighed, combined with 2 g of sodium bicarbonate, diluted to 1 L with distilled water, and carefully shaken until completely dissolved. After that, the medium was filtered through a Millipore bacterial filter to remove any contaminants (0.22 mL). The prepared medium was kept in a refrigerator (4 °C) and inspected for contamination at regular intervals. The medium was warmed in a water bath to 37 °C before use and supplemented with penicillin–streptomycin and FBS. The RPMI-1640 medium was prepared with sodium bicarbonate. For feasibility counting, an isotonic trypan blue solution (0.05%) was prepared in normal saline. Prior to use, the RPMI-1640 medium was supplemented with 10% FBS (heat inactivated at 56 °C for 30 min), 100 units/mL penicillin, and 2 mg/mL streptomycin. Picking cells was done with trypsin (0.25 × 10−1% w/v). Unbound SRB dye was dissolved in acetic acid (1% v/v). As a protein dye, SRB (0.40%) dissolved in 1% acetic acid was used. A 50% trichloroacetic acid stock solution was prepared and stored. For protein precipitation, 50 µL of the stock was added to 200 µL of RPMI-1640 medium per well, resulting in a final concentration of 10%. The solvents used were 100% isopropanol and 70% ethanol. For SRB dye solubilization, Tris base (10 mM; pH = 10.50) was used. The pH was adjusted with hydrochloric acid after the tris base (121.10 g) was dissolved in 1000 ml of distilled water (2 M).
Instrumentation
Microanalyses of carbon, hydrogen, and nitrogen were carried out at the Microanalytical Center, Cairo University, Egypt, using a CHNS-932 (LECO) Vario elemental analyzer. Analyses of the metals were controlled by dissolving the solid complexes in concentrated HNO3 and dissolving the remainder in deionized water. The metal content was carried out using inductively coupled plasma atomic absorption spectrometry (ICP-AES), Egyptian Petroleum Research Institute. Fourier transform infrared (FT-IR) spectra were recorded with a Perkin Elmer 1650 spectrophotometer (400–4000 cm−1) as KBr pellets. 1H NMR spectra, as solutions in DMSO-d6, were chronicled with a 300 MHz Varian-Oxford Mercury at room temperature using tetramethylsilane as an interior standard. Mass spectra were chronicled using the electron ionization technique at 70 eV with an MS-5988 GS–MS Hewlett-Packard instrument at the Microanalytical Center, National Center for Research, Egypt. UV–Vis spectra were acquired with a Shimadzu UVmini-1240 spectrophotometer. Molar conductivities of 10−3 M solutions of the solid complexes in DMF solvent were measured with a Jenway 4010 conductivity meter. Thermogravimetric (TG) and differential thermogravimetric (DTG) analyses of the solid complexes were carried out from room temperature to 1000 °C using a Shimadzu TG-50H thermal analyzer. Antimicrobial measurements were carried out at the Microanalytical Center, Cairo University, Egypt. Anticancer activity experiments were performed at the National Cancer Institute, Cancer Biology Department, Pharmacology Department, Cairo University. The optical density (OD) of each in good form was measured spectrophotometrically at 564 nm with an ELIZA microplate reader (Meter tech. R960, USA) The surface area of substances, as well as gas adsorption measurements, was carried out at 77 K with N2 as the adsorptive gas. The Brunauer–Emmett–Teller (BET) theory was used to measure the analysis, which was done with a Nova Touch LX2 analyzer.
Synthesis of Schiff base ligand
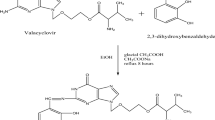
The symmetric Schiff base ligand (L) was made by condensation of 2,2-(ethylenedioxy)bis(ethylamine) and imidazole-2-carboxaldehyde in a 1:2 molar mixture. Dropwise additions of 2,2-(ethylenedioxy)bis(ethylamine) (15.6 mmol, 3 mL) to imidazole-2-carboxaldehyde (31.22 mmol, 3 g) dissolved in DMF were produced. The resulting mixture was stirred in a reflux bath for 4–5 h, and the brown solid compound was separated. It was desiccated in vacuum after being filtered, recrystallized, washed with diethyl ether, and filtered again (Scheme 1).
Synthesis of metal complexes
The complexes of Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), and Cd(II) metal ions were made by reacting a 1:1 molar mixture of a hot ethanolic solution (60 °C) of the appropriate metal chloride (0.98 mmol) with Schiff base ligand (L; 0.3 g, 0.98 mmol). The complexes were precipitated after 1 h stirring under reflux. Filtration was used to extract them, and diethyl ether was used to purify them many times.
Pharmacology
Antibacterial activities
Antimicrobial activity of the tested samples was determined by a modified Kirby-Bauer disk diffusion method [12]. Briefly, 100 µL of the test bacteria was grown in 10 mL of fresh media until they reached a count of approximately 108 cells/mL for bacteria. 100 µL of microbial suspension was spread onto agar plates corresponding to the broth in which they were maintained. Isolated colonies of each organism that may be playing a pathogenic part should be selected from primary agar plates and tested for susceptibility by disk diffusion method. Plates inoculated Gram (+) bacteria as Staphylococcus aureus and Bacillus subtilis; Gram (−) bacteria as Escherichia coli and Pseudomonas aeruginosa. They were incubated at 35–37 °C for 24–48 h, and then, the diameters of the inhibition zones were calculated in millimeters.
Standard disks of amikacin (antibacterial agent), served as positive controls for antimicrobial activity, but filter disks impregnated with 10 µL of solvent (distilled water, chloroform, DMSO) were used as a negative control. Blank paper disks (Schleicher & Schuell, Spain) with a diameter of 8.0 mm were impregnated 10 µL of tested concentration of the stock solutions (20 mg/mL). When a filter paper disk impregnated with a tested chemical is placed on agar, the chemical will diffuse from the disk into the agar. This diffusion will place the chemical in the agar only around the disk. The solubility of the chemical and its molecular size will determine the size of the area of chemical infiltration around the disk. If an organism is placed on the agar it will not grow in the area around the disk if it is susceptible to the chemical. This area of no growth around the disk is known as a “Zone of inhibition” or “Clear zone.” For the disk diffusion, the zone diameters were measured with slipping calipers of the National Committee for Clinical Laboratory Standards. Agar-based methods such as E-test and disk diffusion can be good alternatives because they are easier and more rapidly than broth-based methods [13].
Anticancer activity
Skehan and Storeng’s [13] approach was used to assess the compounds’ potential cytotoxicity. To allow cell attachment to the plate wall, cells were plated in a 96-multiwell plate (104 cells/well) for 24 h before treatment with the compounds. Different concentrations of the compounds under investigation (0, 5, 12.5, 25, 50, and 100 μg/mL) were applied to the cell monolayer. The monolayer cells were kept warm with the compounds for 48 h at 37 °C in a 5% CO2 atmosphere. Cells were immobilized, cleaned, and stained with SRB stain after 48 h. The excess stain was removed with acetic acid, and the attached stain was revived with tris–EDTA buffer. Using an ELIZA microplate reader, the optical density (O.D.) of each well was determined spectrophotometrically at 564 nm, the mean background absorbance was automatically subtracted, and mean drug concentrations were estimated. The survival curve of a breast tumor cell line is plotted using the relationship between drug concentration and surviving fraction. The following formula was used to measure the percentage of cells that survived:
The IC50 values (the concentrations of the Schiff base ligand (L) or its metal complexes were required to produce 50% inhibition of cell growth) [14].
Computational methodology
The Gaussian09 software suite was created to calculate the electronic structure of the Schiff base ligand (L) and Ni(II) complex. For complete optimization, the DFT-based B3LYP approach was used, along with the LANL2DZ basis regulate. The TD-DFT method (along with the LANL2DZ basic set) was used to measure the electronic absorption spectra of the ligand and its Ni(II) complex in order to account for the influence of the solvent around the molecule.
Molecular docking
To determine the best active compound binding modes against the crystal structure of the SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19), researchers used the crystal structure of the SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) (PDB ID: 6Y84). (9) MOE2008 software was used to conduct molecular docking tests. It is a rigid molecular docking software and an interactive molecular graphics application for measuring and showing possible receptor and ligand docking modes, as well as complex molecules. It requires the ligand and the receptor as input in PDB format. The amino acid chain was kept and the water molecules and crystallized ligands and chloride ions in out sphere were removed. The structure of ligand in PDB file format was created using Gaussian09 software. Crystal structure of the SARS-CoV-2 (COVID-19) was downloaded from the protein data bank (http://www.rcsb.org/pdb) [15].
Results and discussion
Characterization of Schiff base
Elemental analyses, mass, IR, and 1HNMR spectroscopy were carried out on the synthesized Schiff base (Fig. 1a). Table 1 summarizes the findings of the elemental tests as well as the melting point. The obtained results are in good agreement with the measured values, and the melting point of the prepared Schiff base is sharp, indicating purity [16].
Composition and structures of Schiff base and its complexes
Elemental analysis, infrared, and electronic spectral methods (UV–Vis spectroscopy), molar conductivity, and magnetic susceptibility measurements were all used, with the latter being the most sensitive and informative [17].
Elemental analysis
The elemental analysis of a compound allows one to establish the compound analytical formula. The empirical formula for a compound is the formula that includes the smallest collection of integer ratios for the elements in the compound that results in the correct elemental composition by mass. As a result, physical properties and elemental analysis of the compounds (C, H, and N) are used to determine the composition of complexes. The found percentages of C, H, and N agree with the suggested composition for the most complexes (Table1) [17].
Infrared spectra
FT-IR spectrum (Table 2) of Schiff base ligand showed a lack of the NH2 stretching band characteristic of 2,2-(ethylenedioxy)bis(ethylamine). On the other hand, a new strong band at 1660 cm−1 in the IR spectrum was assigned to the azomethine nitrogen of the ligand. This band was shifted to lower frequency in the corresponding metal complexes of that ligand [18]. The azomethine band was found in the IR spectra of Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), and Cd(II) complexes at 1623, 1636, 1630, 1649, 1633, 1631, and 1648 cm−1, respectively [19]. This shift pointed out the binding of azomethine N to metal ions. This was further confirmed by the appearance of bands at around 420–471 cm−1, corresponding to the ν(M–N) stretching vibration [20].
Two strong bands observed in the IR spectrum of the ligand at 1568 and 1454 cm−1 which may be assigned to the combination bands (NH) and (C=N) bending vibrations of imidazole ring, respectively. These bands were not shifted in the spectra of the complexes, indicating that the imidazole N is not involved in the coordination [21].
FT-IR spectra of the complexes also showed strong bands in the 3422–3430 cm−1 region, suggesting the presence of water in the complexes. The coordinated water molecules generally showed characteristic peaks at 872–748 cm−1 due to ρr(H2O) and at 784–672 cm−1 ρw(H2O) except Ni(II) and Cd(II) complexes. The infrared spectra showed stretching frequencies in the region 528–612 cm−1, corresponding to υ(M–O) vibration. In the free Schiff base ligand, the band at 1118 cm−1 due to etheric υ(C–O) was shifted to lower wavenumber in the complexes indicating the coordination of the etheric oxygen atom to the metal ions. Therefore, it can be concluded that coordination took place via etheric oxygen and azomethine nitrogen of the Schiff base ligand molecule [22,23,24].
1H NMR Spectrum
The 1H NMR spectrum of the ligand showed a singlet signal at 8.32 ppm, which corresponds to the azomethine proton. Upon examinations, it is found that azomethine proton of Schiff base ligand was shifted up field in the Cd(II) complex at δ 9.59 ppm. This up-field shifting of azomethine proton in Cd(II) complex was attributed to the discharging of electronic cloud toward the Cd(II) ion indicating coordination through the azomethine nitrogen to the metallic ion. The signal appeared at 4.91 ppm due to NH imidazole ring proton [19], which disappeared upon adding deuterated solvent confirming its position. The position of NH proton still in the same position (4.91 ppm) in the Cd(II) complex. This means that the imidazole NH does not involve in coordination [19]. Doublet signals at 7.13 and 8.17 ppm were assigned for –CH protons of imidazole ring. In the spectrum of Cd(II) complex, doublet signals at 7.11 and 7.99 ppm were assigned for –CH protons of imidazole ring. The absence of peak at around 3.5 ppm assigned to –NH2 group indicated desired product formation [25]. The multiple signals from 2.51 to 3.71 ppm correspond to CH2 protons of ethylene moiety in the free Schiff base ligand [20]. These signals were found in the spectrum of Cd(II) complex at δ = 2.49–3.58 ppm, corresponding to CH2 protons of ethylene moiety [16, 20, 22]. The data of the 1H NMR and 13CNMR spectra of Schiff base ligand (L) and its mononuclear Cd(II) complex were shown in Supplementary Fig. 1S, 2S.
Mass spectra and molar conductivity
The molecular ion peak appears in the mass spectrum of the ligand at (m/z = 305.10), which corresponds to its formula weight (305.29). The molecular ion peak appears in the mass spectral values of the cobalt complex at (m/z = 561.40), which is consistent with its formula weight (561.00). The spectrum was also revealed a molecular ion peak at 305.00 amu, which corresponds to the Schiff base ligand and confirms its metal ion binding (Fig. 2).
To determine the ionic nature of the metal complexes, the molar conductivity of all metal complexes in DMF solution (10−3 M) was calculated. Except for Ni(II) and Cd(II) complexes, the high molar conductance values suggested that remaining metal complexes were electrolytes (Table 1) [19].
Electronic spectra and magnetic studies
To determine the structure of complexes, magnetic susceptibility measurements and electronic spectral data have been getting through. The efficient magnetic moment (eff) values for complexes with electronic spectra of ligand and its metal complexes have been observed at room temperature (300 K). On wavelengths ranging from 200 to 800 nm, the electronic spectra of ligand and its metal complexes in DMSO solutions were recorded.
The electronic spectrum of free Schiff base ligand shows a broad band at 291 and 340 nm, which are assigned to π–π* transition of the C=N chromophore and n–π* transition, respectively [26]. The spectra of complexes showed peaks in range 339–361 cm−1and 285–307 nm related to n–π* and π–π* transitions, respectively [27].
Chromium(III) complex showed magnetic moment resembling to three unpaired electrons, i.e., 4.03 B.M., expected for high-spin octahedral chromium(III) complexes. Six coordinated Cr(III) complexes with octahedral expose three spin allowed bands within the range of 18,000–30,000 cm−1 [27]. The Cr(III) complex displayed bands at 17,513, 25,000, 28,248, and 34,843 cm−1, which may be assigned to 4B1g → 4Eag, 4B1g → 4B2g, 4B1g → 4Ebg, and 4B1g → 4A1g transitions, respectively [27, 28].
Mn(II) complex showed magnetic moment value of 5.71 BM which is very close to the theoretically calculated (6.0 BM) value [14, 29]. This suggested an octahedral environment around the Mn(II) ion [30]. The complex below study showed three bands in the range of 17,006–25,510 cm−1. The Mn(II) complex displayed bands at 17,006, 23,256, and 25,510 cm−1, which may be appointed to 6A1g → 4T1g(G); 6A1g → 4T2g(G) and 6A1g → 4T1g(P) transitions, respectively. Thus, the ligand field bands and magnetic moment value support an octahedral geometry around Mn(II) [14, 29].
Iron (III) complex viewed magnetic moments corresponding to three unpaired electrons, i.e., 5.32 B.M., usual for high-spin octahedral Iron(III) complexes [14]. Six coordinated Fe(III). The Fe(III) complex displayed bands at 16,835, 17,301, and 21,739 cm−1, that may be assigned to 4T2g(G) → 6A1g, 4T2g(G) → 6A1g, and 4T1g(D) → 6A1g transitions, respectively, indicating the octahedral geometry of the complex [14].
The electronic spectrum of Co(II) complex exposed three broad peaks at 15,198, 16,420, and 23,041 cm−1 assigned to 4T1g → 4T2g(F), 4T1g → 4A2g(F), and 4T1g → 4T1g(P) transitions, respectively. The magnetic moment value of the Co(II) (d7) complex is (5.06 B.M). The spectrum seems like those reported for octahedral complex [14, 28, 31].
Nickel (II) complex gave magnetic moment value of 3.32 B.M, which falls within the range of octahedral Ni(II) complexes. The diffused reflectance spectrum of this complex showed three absorption bands at 17,637, 18,622, and 22,271 cm−1. These bands may be given to 3A2g → 3T2g(F), 3A2g → 3T1g(F) and 3A2g → 3T1g(p) transitions, respectively, inside an octahedral environment. This indicates an octahedral geometry as the results of the presence of two unpaired electrons and high-spin sp3d2 hybridization [31, 32].
The diffused reflectance spectrum of the copper(II) complex exposes one d-d absorption band at 22,624 cm−1. This band is given to 2T2g → 2Eg (D) transition. The magnetic moment value of the Cu(II) complex was found to be 1.98 B.M which designates the existence of Cu(II) complex in octahedral structure [33, 34].
Generally, Cd(II) complex did not expose any d–d electronic transition because of its completely filled d10 and it proposed to have octahedral structure [14, 26].
Thermogravimetric analysis
The decomposition behavior of the Schiff base ligand and its new complexes was determined using thermogravimetric analysis (Table 3). Also, it was used to determine if the water molecules (if any are available) are outside or within the central metal ion’s inner sphere coordination.
The Schiff base ligand with the molecular formula (C14H21N6O2) was thermally decomposed in two successive decomposition steps with a first and second estimated mass losses of 99.87% (calcd. 100%) at 30–900 °C, which may be recognized to the decomposition of the Schiff base ligand (C14H21N6O2 molecule) into gases.
The thermogram of Cr(III) chelate exposed seven decomposition steps within the temperature range from 30 to 900 °C. The first three steps of decomposition within the temperature range from 30 to 270 °C correspond to the loss of 4H2O and 1.5Cl2 gases with an estimated mass loss of 34.30% (calcd. 33.30%), while the final four steps occur within the temperature range from 270 to 900 °C and correspond to the removal of C10H21N6O0.5 molecule as gases with a mass loss of 43.08% (calcd. 43.51%). By the removal of the organic part of the ligand, metal oxide contaminated by carbon atoms as a residue was left. The total weight loss amounted to 77.38% (calcd. 76.81%).
The TG curve of the Mn(II) chelate showed three decomposition steps within the temperature range from 30 to 900 °C. The three steps of decomposition correspond to the loss of Cl2, 4H2O and C9H21N6O molecules as gases with a mass loss of 74.98% (calcd. 73.97%). By the removal of the organic part of the ligand, metal oxide contaminated by carbon atoms was remained as a residue. The overall weight loss amounted to 74.98% (calcd. 73.97%).
The TG curve of the Fe(III) chelate showed three decomposition steps within the temperature range of 30–900 °C. The three steps of decomposition correspond to the loss of 1.5Cl2, 2H2O and C12H21N6O0.5 molecules as gases with a mass loss of 79.53% (calcd. 79.39%). Metal oxide contaminated by carbon atoms was remained as a residue after the removal of the organic part of the ligand. The overall weight loss amounted to 79.53% (calcd. 79.39%).
The TG curve of the Co(II) and Ni(II) chelates showed three decomposition steps within the temperature range from 30 to 900 °C. The first step of decomposition within the temperature range from 30 to 121 °C and from 30 to 151 °C corresponds to the loss of 3H2O molecules with an estimated mass loss of 9.08% (calcd 9.63%) and 11.53% (calcd 11.04%) for Co(II) and Ni(II) chelates, respectively. The second step of decomposition within the temperature range from 121 to 285 °C corresponds to the loss of 4H2O molecules with an estimated mass loss of 12.80% (calcd 12.83%) and from 151 to 305 °C corresponds to the loss of Cl2 gas with an estimated mass loss of 14.71% (calcd 14.52%) for Co(II) and Ni(II) chelates, respectively. In addition, the final step occurred within the temperature range from 285 to 900 °C and from 305 to 900 °C corresponds to the removal of Cl2 and C9H21N6O molecules and C10H21N6O molecule as gases with a mass loss of 52.37% (calcd. 53.48%) and 48.92% (calcd 49.28%) for Co(II) and Ni(II) chelates, respectively. By the removal of the organic part of the ligand, metal oxide contaminated by carbon atoms was left as a residue. The overall weight loss amounted to 74.25% (calcd 75.94%) and 75.16% (calcd 74.84%) for Co(II) and Ni(II) complexes, respectively.
The thermogram of Cu(II) and Cd(II) chelates showed three decomposition steps within the temperature range from 30 to 900 °C. The first two steps of decomposition within the temperature range from 30 to 207 °C and from 30 to 309 °C correspond to the loss of 4H2O and 0.5Cl2 gases and loss of 2H2O molecules and Cl2 gases with an estimated mass loss of 20.43% (calcd 21.02%) and 21.61% (calcd 20.40%) for Cu(II) and Cd(II) complexes, respectively, while the final step occurred within the temperature range from 207 to 900 °C and from 309 to 900 °C corresponds to the removal of 1/2Cl2 and C10H21N6O molecule and removal of C14H21N6O molecule as gases with a mass loss of 54.70% (calcd 54.06%) and 55.05% (calcd 55.11%) for Cu(II) and Cd(II) complexes, respectively. By the removal of the organic part of the ligand, metal oxide contaminated by carbon atoms was left as a residue. The overall weight loss amounted to 75.13% (calcd 75.08%) and 76.66% (calcd 75.51%) for Cu(II) and Cd(II) complexes, respectively.
BET surface area study
The BET measurements (Table 4) were carried out to calculate the surface area of the metal complexes which showed high surface area in the range of 121–298 m2 g−1. It is to be observed that the high surface area could be contributed to the mesoporous structure and nano-size of the particles which is in the range of 1.13–6.66 nm [35].
Surface area and pore volume values approximated by nitrogen adsorption isotherms at relative pressures (p/pº) are given in Table 4. The micropore volumes and SBET values of entrapped complexes showed a range pore volume (0.22–0.09 cc/g) and singlet surface area (135–48 m2/g) [31].
Structural interpretation
The structures of the tetradentate Schiff base ligand (L) with Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), and Cd(II) metal ions were characterized with elemental analyses, molar conductivity, IR,1H-NMR, UV–Vis, mass and thermal analyses; then, the recommended structures of transition metal complexes were reported in Fig. 3
Geometry optimization
The fully optimized geometries of the ligand and its Ni(II) complex were shown in Fig. 1. The values of the selected bond lengths and bond angles calculated for Ni(II) complex showed octahedral geometry around the Ni(II) ion (Table 5). A slight elongation in bond lengths of C29–N32, N11–C1, C15–O18, and C26–O25 was noted in Ni(II) complex as ligand coordinated via azomethine nitrogen and etheric oxygen. The O(6)–Ni(42), O(12)–Ni(42), N(19)–Ni(42), N(31)–Ni(42), Cl(43)–Ni(42), and Cl(42)–Ni(44) bond lengths were found to be 1.84, 1.85,1.85, 1.86, 2.14, and 2.14 A°, respectively. Two azomethine nitrogens and two etheric oxygens occupied two of the four positions, while the chloride ion occupied the remaining two. As previously mentioned, the bond angles in the coordination sphere of the Ni(II) complex suggested an octahedral geometry. Intramolecular hydrogen bonds may be accounted for the decrease in metal-chloride angles [14].
Molecular electrostatic potential (MEP)
Electrostatic potential V(r) maps were used to research the coordination sites present in the Schiff base ligand since they are known for identifying the electronic charge distribution across molecular surfaces and, as a result, predicting reaction sites. The same basis set that was used for optimization was used to create these maps. In this analysis, 3D MEP plots for the ligand and its Ni(II) complex were drawn. Based on the MEP, the electron-rich region (red on the map) can be ordered in general (favor site for electrophilic attack). The electron-poor field, on the other hand, is blue (favor site for nucleophilic attack). The green-colored region, on the other hand, denotes the neutral electrostatic potential region. It can be shown that the Schiff base ligand (L) is stable, with a nearly uniform charge density distribution. However, the negative charge surface surrounding oxygen and nitrogen atoms is greater, making these sites potentially more suitable for electrophilic attack (red color) (Fig. 4).
In terms of electron density, the aromatic ring appears to be neutral. Thus, the potential distribution favors the complexation reaction, as shown by the electrostatic potential distribution of the Ni(II) complex, which has a greater negative charge surrounding the metal core (Fig. 4b). The Mulliken electronegativity also showed that the electronegativity of oxygen and nitrogen in Ni(II) complex was higher than that of free Schiff base ligand (L), indicating that they are more susceptible to electrophilic attack by metal ion [14].
Molecular parameters
Additional parameters such as the highest occupied molecular orbital energy (EHOMO), the lowest unoccupied molecular orbital energy (ELUMO), ∆E, absolute electronegativities, χ, chemical potentials, Pi, absolute hardness, η, absolute softness, σ, global electrophilicity, ω, global softness, S, and additional electronic charge, ∆Nmax have been estimated for the free Schiff base ligand and its Ni(II) complex and listed in Table 5. Electrophilicity index (ω) is one of the most important quantum chemical descriptors in describing toxicity and the reactivity of various selective sites. The electrophilicity may quantify the biological activity of drug receptor interaction. Also, this index measures the stabilization energy when the system acquires extra negative charge from the environment. η and σ indexes are the measure of the molecular stability and reactivity; also, their concepts are related to each other. The softness indexes are the vice versa image for global hardness. These parameters are useful to support the suggested structures. The mentioned quantum chemical parameters were calculated with the help of the following equations:
The data calculated are presented in Table 5 and reflected the following notes:
(1) The data of Schiff base ligand and its Ni(II) complex had a great chance and priority for biological activity based on high ω value.
(2) S and ω were the softness indexes, while η are for hardness indication; a hard molecule had a high stability due to its high energy difference in-between the EHOMO and ELUMO than the soft molecule. So, the soft molecule was the reactive one having flexible donation toward the metal ions. Accordingly, the investigated Schiff base ligand molecule was soft toward the coordination.
(3) The positive electrophilicity index (χ) value and the negative electronic chemical potential (μ) value indicated that the Schiff base ligand molecule capable of accepting electrons from the environment and its energy must decrease upon accepting electronic charge. Therefore, the electronic chemical potential must be negative.
(4) The Ni(II) complex showed high values of dipole moments than the free ligand.
Both the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) were the main orbitals that participate in chemical stability. The HOMO represents the ability to donate an electron; LUMO as an electron acceptor represents the ability to obtain an electron. From the attained data (Table 5), it can assume that:
-
1.
The energies of the HOMO and LUMO were negative values and more negative than free Schiff base ligand, which showed the stability of isolated complex [14].
-
2.
The EHOMO and ELUMO values of Ni(II) complex were calculated and showed a decrease than the free Schiff base ligand which represents the strength of M–L shorter bonds.
-
3.
The total energy of Ni(II) complex was higher than free ligand, which indicated greatly the stability of the isolated solid complex.
-
4.
The small energy gap can be associated with a high chemical reactivity, low kinetic stability, and reflects to efficient electronic charge transfer interaction making the molecule highly polarizable.
-
5.
The HOMO level was mostly localized on the azomethine nitrogen and oxygen of phenol group in the ligand, which indicated the preferable sites for nucleophilic attack to the central metal atom [14].
UV–Vis spectra
Understanding the photochemistry of transition metal compounds necessitates an understanding of molecular orbital properties as well as sufficient excited states. Since they govern electronic excitations and transition characters, frontier orbitals played an important role in such systems. It is possible to make comments about the contributions of the ligand and metal orbitals to molecular orbitals using TD-DFT calculations. Since it is impractical to study all the electronic transitions and orbitals, some limitations were imposed. TD-DFT has become famous for theoretical investigation of electronic spectra of molecules due to its high accuracy and low computational cost. At the TD-DFT/B3LYP/LANL2DZ level of theory, thirty singlet states were investigated for low-lying excited states on optimized ground state structures of Schiff base ligand and its Ni(II) complex. In Table 6, the experimental and theoretical electronic spectra were presented. The TD-DFT calculations have been evaluated in the N,N-dimethylformamide solvent background and compared with the experimental data. The transitions between interferometer orbitals for wavelengths corresponding to maximum oscillator strength of simulated results with contemporary experimental observations are presented in Fig. 5a, b. For example, the electronic transitions for Schiff base ligand obtained at calculated 288 nm correspond to experimental peak at 291 nm. This transition has been majorly contributed from HOMO-2 to LUMO transitions which was primarily π → π* in nature. The different transitions and its experimental counterpart of the free ligand and its Ni(II) complex are summarized in Table 6 [14].
Antimicrobial assays
Antibacterial studies
The ligand and its metal complexes were tested for antibacterial activity against human pathogens such as S. aureus and E. coli. Table 7 summarizes the findings. The antibacterial activity of the ligand and its complexes was found to be effective against S. aureus and E. coli, except for the ligand and Cr(III) complex with S. aureus. As a result of the chelating effect, metal complexes become powerful antibacterial agents, inhibiting, or destroying microorganisms by blocking their active sites [6].
Antifungal studies
The ligand and its Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), and Cd(II) complexes have been tested on behalf of their antifungal activity. The results are exposed in Table 7 and Fig. 6. The results showed that Cd(II) complex exhibits enhanced activity against Aspergillus flavus and Ni(II) complex against Candida albicans than the standard drug ketoconazole [6].
The activities of the prepared Schiff base ligand and its metal complexes were approved via calculating the activity index according to the following relation and that exposed in Fig. 7a and b: activity index (A) = inhibition zone of compound mm/inhibition zone of standard drug mm × 100.
Even though the exact mechanism is not understood biochemically, mode of action of antimicrobial may involve various targets in microorganisms.
-
1.
Interference with the cell wall synthesis, damage because of which cell permeability may be altered (or) they may disorganize the lipoprotein leading to the cell death.
-
2.
Deactivate different cellular enzymes, which play a critical role in different metabolic pathways of these microorganisms.
-
3.
Denaturation of one or more proteins of the cell, because of which the normal cellular processes are impaired.
-
4.
Formation of a hydrogen bond between the active center of cell constituents and the azomethine group, interfering with the normal cell operation. The higher inhibition of microbial growth is due to uncoordinated hetero atoms moieties, as shown in Table 7. The ligand (L) in the complexes had some uncoordinated donor atoms that enhanced the complexes’ operation by bonding with trace elements found in microorganisms. This could combine with the uncoordinated site and prevent microorganisms from growing [18, 36].
To assess the biological profile, another series of antimicrobial agents based on metals with a [(1E)-1-(2-hydroxyphenyl)ethylidene]-2-oxo-2H-chromene-3-carbohydrazide Schiff base possessing a NOO donor system was synthesized and thoroughly analyzed. Cd(II) complex showed the maximum zone of inhibition (18 and 16 mm) against E. coli and A. flavus organisms compared to [Cd(L)Cl2]2H2O complex which showed the maximum zone of inhibition (27 and 31 mm) [37, 38]. Co(II) complex of the same SB showed the maximum zone of inhibition (12 mm) against Staphylococcus aureus compared to [Co(L)H2OCl]Cl·6H2O complex which showed the maximum zone of inhibition (20 mm) [38].
Also, another series of antimicrobial agents based on metals with 2-(((1,5-dimethyl3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)imino) (phenyl)methyl)benzoic acid Schiff base, Ni(II) complex showed the maximum zone of inhibition (18 mm) against C. albicans compared to [Ni(L)Cl2]3H2O complex which showed the maximum zone of inhibition (23 mm) [39, 40].
It can be pointed out from these results that the reported complexes have higher antimicrobial activity that the previously reported complexes as indicated from the inhibition zone.
Formation of a hydrogen bond between the active center of cell constituents and the azomethine group, interfering with the normal cell operation. The higher inhibition of microbial growth is due to uncoordinated hetero atoms moieties, as shown in Table 7. The Schiff base ligand (L) in the complexes had some uncoordinated donor atoms that enhanced the complexes’ operation by bonding with trace elements found in microorganisms. This could combine with the uncoordinated site and prevent microorganisms from growing [37].
Such increase activity of the metal complexes can be explained based on Overtone’s concept and Tweedy’s chelation theory. There is a decrease in the polarity of the metal atom significantly after chelation, because of the partial sharing of its positive charge with the donor groups and due to π-electron delocalization overall chelate ring [41].
Anticancer effects of Schiff base and its complexes
The Schiff base and its complexes have been tested for their anticancer effect to the human breast cancer cell line (MCF-7) with various concentrations. The anticancer activity results of the compounds at 100 µg/mL were 67, 67, 70, 48, 54, 60, 57, and 56 for ligand and its Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), and Cd(II) complexes, respectively, then choose the highest activity and prepared four concentration as shown in Table 8. % cell inhibition and IC50 values were found out and indicate that Cu(II) complex had higher sensitivity toward the breast cancer line than the free ligand and other complexes as shown in Fig. 8 [42].
The antitumor activity against MCF-7 breast cancer cell lines was tested in vitro. The results are shown in Table 6. IC50 values obtained according to the corresponding formula are shown in Table 6. Comparing the IC50 values of ligand, Cr(III) and Mn(II) complexes, it can be found that the Mn(II) complex had stronger activity against human breast cancer cells (MCF-7) than the other complexes, and its IC50 value was 21 mg/L, and the IC50 value of cisplatin was 14.52 mg/L, the best anti-human breast cancer cell MCF-7 activity. Currently, the IC50 value of cisplatin is more than 100 mg/L. These results provide more possibilities for the research and development of metal anticancer drugs and provide some experimental data for the development and application of metal complexes anticancer drugs, but their performance should be further studied [43].
Molecular modeling: docking study
Molecular docking is a valuable technique to predict the stable structure of receptor–ligand complex for better recognition of the interaction details in drug discovery process. This method is frequently employed as virtual searching tools in primary steps of drug design and development [44, 45].
The molecular docking studies were undertaken to closely examine the interaction between the synthesized compounds and crystal structure of the SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) (PDB ID: 6Y84) proteins. The results in Table 9 showed that the compounds interacted favorably with the active binding sites of the proteins.
When we analyzed the out of 8 predicted binding conformations of ligand, Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), and Cd(II) with 6Y84 docking, the best confirmations exhibited that the free energies of binding (ΔG) were − 6.0, − 7.6, − 10.1, − 8.5, − 5.9, − 7.7, − 4.6, and − 6.1 kcal/mol, respectively. All Schiff base complexes are binding through H-donor except ligand is binding by H-acceptor with 3.07, 3.03, 2.99, 2.98, 3.06, 302, 2.95, and 3.07 A° distances, respectively [38, 46, 47].
Strong binding affinities indicate that our compounds can inhibit the biochemical processes of these proteins. [Mn(L) H2O Cl]Cl·3H2O gave the highest binding with the lowest energies (− 10.10 kcal/mol). This corroborated well with the antioxidant result in Fig. 9 The binding mode of the Mn(II) complex with the target protein is shown in Fig. 10 [47].
Conclusion
The structures of the synthesized Schiff base ligand and its Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), and Cd(II) complexes were characterized based on analytical and spectroscopic data. Experimental results indicated the approval of octahedral geometry for the metal(II)/(III) complexes and the participation of the Schiff base ligand in chelation in a tetradentate fashion. Also, structural characterization showed big surface area and small pore size according to the BET results. It can be used in different nanotechnology applications. Metal(II)/(III) complexes exhibited better antibacterial and antifungal activities than the parent Schiff base ligand in the identical experimental conditions. The increased activity of the metal complexes can be rationalized based on chelation theory. Chelation decreases the polarity of the metal ion because positive charges of the metal are partially shared with the donor atoms extent in the ligands and there may be π-electron delocalization over the whole chelation. This phenomenon improved the lipophilic character of the metal chelate and favored its permeation more efficiently, through the lipoid layer of the microorganism. However, the results of antimicrobial activity are related to their ability to make complexes with soluble extracellular proteins and with the cell wall, for the lipophilic character of these compounds, which may bring about the rupture of the cell membrane of microorganisms. In addition, in bacteria, the permeability of the cell membrane is associated with the loss of ions as well as the decrease of its potential, causing damage that may lead to the extravasation of macromolecules, resulting in a collapse of the cellular functions and, consequently, the bacterial death.
The outcomes indicated that all the complexes exhibited better anticancer activity in contrast to MCF-7. Generally, it provides good experimental data and theoretical guidance aimed at the progress and application as anticancer drugs in the future; however, its anticancer mechanism and other characteristics need to be further studied. Ligand was effective to fight against the new coronavirus. Its Mn(II) complex with a lower binding energy than the ligand (L) implies a possible strong antiviral activity. This study will be conducted to be tested for the treatment against new coronavirus.
References
K.L. Barry, C.D. Grimmer, O.Q. Munro, M.P. Akerman, RSC Adv. 10, 7867 (2020)
P. Martins, J. Jesus, S. Santos, L.R. Raposo, C. Roma-Rodrigues, P.V. Baptista, A.R. Fernandes, Molecules 20, 1685 (2015)
H. Kargar, F.A. Meybodi, R.B. Ardakani, M.R. Elahifard, V. Torabi, M.F. Mehrjardi, M.N. Tahir, M. Ashfaq, K.S. Munaware, J. Mol. Struct. 1230, 129908 (2021)
K.L. Barry, C.D. Grimmer, O.Q. Munro, M.P. Akerman, RCS Adv. 10, 7867 (2020)
H. Karga, R.B. Ardakani, V. Torabi, A. Sarvian, Z. Kazemi, Z.C. Natanzi, V. MirkhanI, A. Sahraei, M.N. Tahir, M. Ashfaq, Inorg. Chim. Acta 514, 120004 (2021)
M. Vairalakshmi, R. Princess, R.A.J.A.S. Johnson, Asian J. Pharm. Clin. Res. 12, 206 (2019)
A.A. Ardakani, H. Kargar, N. Feizi, M.N. Tahir, J. Iran. Chem. Soc. 15, 1495 (2018)
H. Kargar, Transit. Met. Chem. 39, 811 (2014)
A. Sahraei, H. Kargar, M. Hakimi, M.N. Tahir, Transit. Met. Chem. 42, 483 (2017)
G. Sivaprakash, P. Tharmaraj, M. Jothibasu, A. Arun, Int. J. Adv. Res. 5, 2656 (2017)
R. Yu, L. Chen, R. Lan, R. Shen, P. Li, Int. J. Antimicrob. Agents 2, 106012 (2020)
A.W. Bauer, W.M. Kirby, J.C. Sherris, M. Turck, Am J Clin Pathol. 45, 493 (1966)
Y.M. Ahmed, W.H. Mahmoud, M.M. Omar, G.G. Mohamed, J. Inorg. Organomet. Polym. Mater. 31, 2339 (2021)
W.H. Mahmoud, M.M. Omar, Y.M. Ahmed, G.G. Mohamed, Appl. Organomet. Chem. 34, 1 (2020)
M. Pal, D. Musib, M. Roy, New J. Chem. 45, 1924 (2021)
H.F. AbdEl-Halim, M.M. Omar, G.G. Mohamed, M.E.E.A. Sayed, Eur. J. Chem. 2, 178 (2011)
A.M. Yimer, Rev. Catal. J. 2, 14 (2015)
R.S. Joseyphus, M.S. Nair, Mycobiology. 36, 93 (2008)
J. Tota, S. Battu, IJPCBS 8, 218 (2018)
P.V. Palaniswamy, M. Dhandapani, J. Suyambulingam, C. Subramanian, J. Serb. Chem. Soc. 85, 215 (2020)
M. Nath, P. Kumar, A. Kumar, J. Organomet. Chem. 695, 1353 (2010)
F.K. Ommenya, E.A. Nyawade, D.M. Andala, J. Kinyua, Hindawi J. Chem. 2020, 1745236 (2020)
M. Shebl, M.A. El-Ghamry, S.M.E. Khalil, M.A.A. Kishk, Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 126, 232 (2014)
M.L. Dianu, A. Kriza, N. Stanica, A.M. Musuc, J. Serb. Chem. Soc. 75, 1515 (2010)
R.R. Surve, S.T. Sankpal, Rasayan J. Chem. 13, 282 (2020)
M.S. Nair, D. Arish, R.S. Joseyphus, J. Saudi Chem. Soc. 16, 83 (2012)
B.H. Al-Zaidi, M.M. Hasson, A.H. Ismail, J. Appl. Pharm. Sci. 9, 45 (2019)
H.H. Bayoumi, A.N. Alaghaz, M.S. Aljahdali, Int. J. Electrochem. Sci. 8, 9399 (2013)
E. Soleimani, J. Chin. Chem. Soc. 57, 653 (2010)
P. Subbaraj, A. Ramu, N. Raman, J. Dharmaraja, J. Saudi Chem. Soc. 19, 207 (2015)
C.K. Modi, P.M. Trivedi, Arab. J. Chem. 10, S1452 (2017)
K.J. Al-adilee, H.K. Dakheel, Eurasian J. Anal. Chem. 5, 13 (2018)
H.I. Alarabi, S.S. Mohamed, W.A. Suayed, I.A. Al-sadawe, M.S. Bensaber, F.M. Sherif, A. Hermann, A. Gbaj, J. Pharm. Pharmacol. Res. 2, 39 (2018)
M.M.H. Khalil, E.H. Ismail, G.G. Mohamed, E.M. Zayed, A. Badr, Open J. Inorg. Chem. 2, 13 (2012)
J.K. Reddy, K. Lalitha, P.V.L. Reddy, G. Sadanandam, M. Subrahmanyam, V.D. Kumari, Catal. Lett. 144, 340 (2014)
C.K. Modi, P.M. Trivedi, Arab. J. Chem. 10, S1452 (2017)
U.D. Kamaci, M. Kamaci, A. Peksel, J. Fluoresc. 27, 805 (2017)
K. Siddappa, K. Mallikarjun, T. Reddy, M. Mallikarjun, C.V. Reddy, M. Tambe, E-J. Chem. 6, 615 (2009)
A. Omanakuttan, G. Priyanka, R.D. Mohan, IOP Conference Series: Materials Science and Engineering (IOP Publishing, Bristol, 2019), p. 50
W.H. Mahmoud, R.G. Deghadi, G.G. Mohamed, J. Therm. Anal. Calorim. 127, 2149 (2017)
A. Sahraei, H. Kargar, M. Hakimi, M.N. Tahir, J. Mol. Struct. 1149, 576 (2017)
A. Palanimurugan, A. Kulandaisamy, J. Organomet. Chem. 861, 263 (2018)
Q. Liu, B. Xie, S. Lin, Q. Liao, R. Deng, Y. Zhaohua, J. Chem. Sci. 131, 1 (2019)
A. Jamshidvand, M. Sahihi, V. Mirkhani, M. Moghadam, I.M. Baltork, S. Tangestaninejad, H.A. Rudbari, H. Kargar, R. Keshavarzi, S. Gharaghani, J. Mol. Liq. 253, 61 (2018)
H. Kargar, R. Behjatmanesh-Ardakani, V. Torabi, M. Kashani, Z. Chavoshpour-Natanzi, Z. Kazemi, V. Mirkhani, A. Sahraei, M.N. Tahir, M. Ashfaq, K.S. Munawar, Polyhedron 195, 114988 (2021)
X. Youdong, X. Meng, Sci. Rep. 10, 1 (2020)
W.H. Mahmoud, R.G. Deghadi, G.G. Mohamed, Appl. Organometal. Chem. 32, e4289 (2018)
Acknowledgements
The authors wish to share their sincere appreciation to Dr. Walaa H. Mahmoud for the revision of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ahmed, Y.M., Omar, M.M. & Mohamed, G.G. Synthesis, spectroscopic characterization, and thermal studies of novel Schiff base complexes: theoretical simulation studies on coronavirus (COVID-19) using molecular docking. J IRAN CHEM SOC 19, 901–919 (2022). https://doi.org/10.1007/s13738-021-02359-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-021-02359-w