Abstract
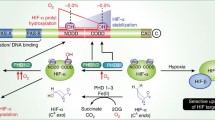
Prolyl hydroxylase domain 2 containing protein (PHD2) is a central protein in regulation of cellular response to hypoxia. This protein controls the responses of cell to oxygen level via the regulation of hypoxia inducible factor (HIF) stability. HIF induces the expression of many genes, especially ones orchestrate angiogenesis. There are some reports that mentioned in some tumor types the level of HIF is high in spite of the presence of wild-type PHD2 and normoxic environment. Therefore, the possibility of PHD2 misfolding in some cancer cells arises. Studying such important protein unfolding pathway is insightful for possible therapeutic approaches. In this study, the unfolding pathway of PHD2 illustrates utilizing molecular dynamics simulation of protein thermal denaturation. Based on current study results, we represent the possible mechanisms of PHD2 unfolding in detail. The possible intermediates of PHD2 thermal unfolding are characterized, and the most venomous state of its unfolding pathway is introduced.








Similar content being viewed by others
References
D.A. Tennant, R.V. Duran, H. Boulahbel, E. Gottlieb, Carcinogenesis 30, 1269–1280 (2009)
K. Takenaga, Front Biosci. 16, 31–48 (2011)
D. Fukumura, R.K. Jain, Microvasc. Res. 74, 72–84 (2007)
P. Nyberg, T. Salo, R. Kalluri, Front Biosci. 13, 6537–6553 (2008)
M. Furuya, Y. Yonemitsu, I. Aoki, Curr. Pharm. Des. 15, 1854–1867 (2009)
D. Liao, R.S. Johnson, Cancer Metastasis Rev. 26, 281–290 (2007)
T. Acker, K.H. Plate, Cancer Treat. Res. 117, 219–248 (2004)
Q. Ke, M. Costa, Mol. Pharmacol. 70, 1469–1480 (2006)
K.A. Lee, J.D. Lynd, S. O’Reilly, M. Kiupel, J.J. McCormick, J.J. LaPres, Mol. Cancer Res. 6, 829–842 (2008)
R.J. Appelhoff, Y.M. Tian, R.R. Raval, H. Turley, A.L. Harris, C.W. Pugh, P.J. Ratcliffe, J.M. Gleadle, J. Biol. Chem. 279, 38458–38465 (2004)
D.A. Chan, A.J. Giaccia, Br. J. Cancer 103, 1–5 (2010)
T. Jokilehto, K. Rantanen, M. Luukkaa, P. Heikkinen, R. Grenman, H. Minn, P. Kronqvist, P.M. Jaakkola, Clin. Cancer Res. 12, 1080–1087 (2006)
H. Zhong, A.M. De Marzo, E. Laughner, M. Lim, D.A. Hilton, D. Zagzag, P. Buechler, W.B. Isaacs, G.L. Semenza, J.W. Simons, Cancer Res. 59, 5830–5835 (1999)
S. Seshadri, K.A. Oberg, V.N. Uversky, Curr. Protein Pept. Sci. 10, 456–463 (2009)
A. Sadana, T. Vo-Dinh, Biotechnol. Appl. Biochem. 33, 7–16 (2001)
F. Ding, J.J. LaRocque, N.V. Dokholyan, J. Biol. Chem. 280, 40235–40240 (2005)
B. Urbanc, L. Cruz, F. Ding, D. Sammond, S. Khare, S.V. Buldyrev, H.E. Stanley, N.V. Dokholyan, Biophys. J. 87, 2310–2321 (2004)
V. Daggett, Methods Mol. Biol. 168, 215–247 (2001)
A. Li, V. Daggett, Proc. Natl. Acad. Sci. U S A 91, 10430–10434 (1994)
V. Daggett, M. Levitt, J. Mol. Biol. 232, 600–619 (1993)
M.W. van der Kamp, V. Daggett, Top. Curr. Chem. 305, 169–197 (2011)
V. Daggett, Acc. Chem. Res. 35, 422–429 (2002)
A.R. Fersht, V. Daggett, Cell 108, 573–582 (2002)
R. Day, B.J. Bennion, S. Ham, V. Daggett, J. Mol. Biol. 322, 189–203 (2002)
J.C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R.D. Skeel, L. Kale, K. Schulten, J. Comput. Chem. 26, 1781–1802 (2005)
M.A. McDonough, V. Li, E. Flashman, R. Chowdhury, C. Mohr, B.M. Lienard, J. Zondlo, N.J. Oldham, I.J. Clifton, J. Lewis, L.A. McNeill, R.J. Kurzeja, K.S. Hewitson, E. Yang, S. Jordan, R.S. Syed, C.J. Schofield, Proc. Natl. Acad. Sci. USA 103, 9814–9819 (2006)
A.D. Mackerell Jr, M. Feig, C.L. Brooks 3rd, J. Comput. Chem. 25, 1400–1415 (2004)
W. Humphrey, A. Dalke, K. Schulten, J. Mol. Graph. 14(33–38), 27–38 (1996)
K. Lindorff-Larsen, S. Piana, R.O. Dror, D.E. Shaw, Science 334, 517–520 (2011)
B. Zagrovic, C.D. Snow, S. Khaliq, M.R. Shirts, V.S. Pande, J. Mol. Biol. 323, 153–164 (2002)
J.K. Myers, C.N. Pace, J.M. Scholtz, Protein Sci. 4, 2138–2148 (1995)
R. Chowdhury, M.A. McDonough, J. Mecinovic, C. Loenarz, E. Flashman, K.S. Hewitson, C. Domene, C.J. Schofield, Structure 17, 981–989 (2009)
S.J. Sheather, C. Jones, J. R. Stat. Soc: Ser. B (Stat. Methodol.) 53, 683–690 (1991)
T.F. Cox, M.A.A. Cox, Multidimensional scaling (Chapman & Hall, London, 1994)
N.A. Baker, D. Sept, S. Joseph, M.J. Holst, J.A. McCammon, Proc. Natl. Acad. Sci. 98, 10037–10041 (2001)
A. Rajan, P.L. Freddolino, K. Schulten, PLoS ONE 5, e9890 (2010)
M.S. Cheung, A.E. Garcia, J.N. Onuchic, PNAS 99, 685–690 (2002)
Y. Levy, J.N. Onuchic, Proc. Natl. Acad. Sci. USA 101, 3325–3326 (2004)
Y.M. Rhee, E.J. Sorin, G. Jayachandran, E. Lindahl, V.S. Pande, Proc. Natl. Acad. Sci. USA 101, 6456–6461 (2004)
Acknowledgments
We gratefully acknowledge the support of the University of Tehran, Iran National Science Foundation (INSF), and the Center of Excellence in Biothermodynamics (CEBiotherm).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
13738_2013_227_MOESM1_ESM.tif
SI-1 To have estimation about the correspondence of PHD2 3D structure and 2D map of X, Y coordinates of PHD2 Cα along 25-ns MD, this graph is generated. Equal regions of 2D map and 3D structure are clarified via arrows. Darker regions are more populated. (TIF 1.48 mb)
13738_2013_227_MOESM2_ESM.gif
SI-2 This animated image shows the distribution of Cl+ ions in simulation space around PHD2 protein along 25-ns MD. PHD2 is represented by ribbon structure. Cl+ ions are presented via color points. Each color of point represents a specific ion with unique atom ID. As this animation demonstrates, a very small fraction of Cl+ atoms penetrated to PHD2 core. (GIF 3.91 mb)
13738_2013_227_MOESM3_ESM.gif
SI-3 This animated image shows the distribution of Na+ ions in simulation space around PHD2 protein along 25-ns MD. PHD2 is represented by ribbon structure. Na+ ions are presented via color points. Each color of point represents a specific ion with unique atom ID. As this animation demonstrates, substantial fraction of Na+ atoms penetrated to PHD2 core. (GIF 7.04 mb)
13738_2013_227_MOESM4_ESM.tif
SI-4 The APBS computed electrostatic potential (EP) of PHD2 is represented. Red surface stands for regions with negative EP. Regions with positive EP are highlighted by blue surface. (TIF 680 kb)
13738_2013_227_MOESM5_ESM.gif
SI-5 The negative electrostatic potential of PHD2 is represented via EP filed lines. This animated image shows that negative EP penetrates into PHD2 lumen, and therefore, such EP attracts positive ions to protein core. (GIF 3.73 mb)
13738_2013_227_MOESM6_ESM.tif
SI-6 This graph represents the interaction energy between ion and PHD2 residue’s types along 25-ns simulation time. Panel (a) shows interaction energies smaller than −0.5 kcal/mol. The (b) and (c) sections represent interaction energies smaller than 1.0 and 2.0 kcal/mol, respectively. The most strong attraction energies between ions and residues present in panel (c). (TIF 1.15 mb)
Rights and permissions
About this article
Cite this article
Hadi-Alijanvand, H., Moosavi-Movahedi, A.A. & Goliaei, B. Unfolding mechanism of PHD2 as a vital protein: all-atom simulation approach. J IRAN CHEM SOC 10, 907–914 (2013). https://doi.org/10.1007/s13738-013-0227-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-013-0227-3