Abstract
Images are frequently used in articles to convey essential information in context with correlated text. However, searching images in a task-specific way poses significant challenges. To minimize limitations of low-level feature representations in content-based image retrieval (CBIR), and to complement text-based search, we propose a multi-modal image search approach that exploits hierarchical organization of modalities and employs both intra and inter-modality fusion techniques. For the CBIR search, several visual features were extracted to represent the images. Modality-specific information was used for similarity fusion and selection of a relevant image subset. Intra-modality fusion of retrieval results was performed by searching images for specific informational elements. Our methods use text extracted from relevant components in a document to create structured representations as “enriched citations” for the text-based search approach. Finally, the multi-modal search consists of a weighted linear combination of similarity scores of independent output results from textual and visual search approaches (inter modality). Search results were evaluated using a standard ImageCLEFmed 2012 evaluation dataset of 300,000 images with associated annotations. We achieved a mean average precision (MAP) score of 0.2533, which is statistically significant, and better in performance (7 % improvement) over comparable results in ImageCLEFmed 2012.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Medical researchers and clinicians routinely use online databases such as MEDLINE® Footnote 1 via PubMed to search for relevant biomedical bibliographic citations. Scientific articles convey information using multiple and distinct modalities, including text and images. For example, authors of journal articles frequently use graphical images (e.g., charts, graphs, maps, diagrams) and natural images (e.g., color or grayscale photographs) to elucidate the text, to illustrate the medical concepts or to highlight special cases. These images often convey essential information in context and can be valuable for improved clinical decision support, research, and education [1, 2].
However, the images contained in biomedical articles are seldom self-evident, and much of the information required for their comprehension can be found in the text of the articles (such as, captions, article titles and abstracts) in which they appear. While images may be separately ambiguous, jointly with co-occurring text they enrich a search response. For example, the authors of a biomedical article [3] report on the size and shape of a nodule depicted in a patient’s lung CT scan as shown in Fig. 1, using the visual characteristics (size = 1 cm and shape = irregular margins) of the region (right middle lobe) as a basis for a possible diagnosis. If an image retrieval system separates this image from its associated caption, as is often done by content-based image retrieval (CBIR) systems [4], it disregards the meaning attached to the appearance of the nodule. In addition, manual annotations are often incomplete and visual properties (such as, peripheral, patchy, ground glass nodular opacities) can often be best described with low level (e.g., color, texture, shape) image features instead of using textual description.
Example image along with its caption in an article [3]
The above example draws the conclusion that single-modality information retrieval, either using text as contextual information or images as visual features, has limitations. Therefore, integration of textual information in a CBIR system or image content information in a text retrieval system might improve retrieval performance [5]. Ingwersen’s cognitive model of information retrieval (IR) [6], which predicts that combining methods using different cognitive structures is likely to be more effective for retrieval than any single method, provides a theoretical basis for the combination of text and images.
Given that images are such a crucial source of information within the biomedical domain, combining textual and visual image features based on multimodal sources has been only recently gaining popularity due to the large amount of information sources. Integration of complementary textual and image information into a unified information retrieval system appears to be promising and could improve retrieval quality through greater utilization of all available (and relevant) information. The results from medical retrieval tracks of previous ImageCLEFFootnote 2 campaigns also suggest that the combination of CBIR and text-based image searches provides better results than using the two different approaches individually [10–12]. While there is a substantial amount of completed and ongoing research in text retrieval, as well as in CBIR in the medical domain, it is not always evident how to exploit the information contained in different sources in an intelligent and task-specific way.
To also enable effective search of diverse images presented in medical journal articles, it might be advantageous for a retrieval system to be able to first recognize the image type (e.g., X-ray, MRI, ultrasound, etc.). A successful categorization of images would greatly enhance the performance of the retrieval system by filtering out irrelevant images, thereby reducing the search space [13]. For example, to search for chest X-rays showing an enlarged heart in a radiographic collection, the database images can first be pre-filtered using automatic categorization by imaging type (e.g., X-ray) and body part (e.g., chest). In addition, the classification information could be utilized to adjust the weights of different image features (such as, color feature could receive more weight for microscopic and photographic images, and edge- or texture-related features for radiographs) in similarity matching for a query and for database images.
Some medical image search engines, such as GoldminerFootnote 3 developed by the American Roentgen Ray Society (ARRS) and YottalookFootnote 4 allow users to limit the search results to a particular modality. However, this modality is typically extracted from the caption and is often not correct or present. Studies have shown that the modality can be extracted from the image itself using visual features, such as the automatic categorization of 6,231 radiological images into 81 categories that is examined [14]; by utilizing a combination of low-level global texture features and a K-nearest-neighbors (KNN) classifier. Recently, a few multimodal classification-based approaches were presented in the ImageCLEFmed medical image classification task. Similar to retrieval, it has shown that the classification results have better accuracy by combining text and images, in most cases, than the results using either text or image features alone [10–12]. However, the various flat classification approaches as described do not exploit the hierarchical structure of image organization at different levels. In addition to using multimodal features, it is possible to exploit the hierarchical organization of the modality classes in order to decompose the task into several smaller classification problems that can be sequentially applied.
Finally, the optimal combination of textual and visual searches should be treated as an information fusion problem. In information retrieval (IR), data fusion is a widely used technique to combine information from various sources to improve the retrieval performance [15, 16]. Many researchers have argued that better retrieval effectiveness may be gained by exploiting multiple query and document representations, retrieval algorithms, or feedback techniques and combining the results of a varied set of techniques or representations.
The majority of the fusion-based retrieval approaches are mainly well studied in the context of the text retrieval domain. Challenges exist in the combination of multimodal information; where the data are often more heterogeneous with multiple example query images with multiple visual (e.g., color, texture, shape, etc.) and textual (e.g., title, caption, mentions, MeSH® Footnote 5, etc.) representations, and collateral text blocks (e.g., caption, mention) have correlations with images. As user query may be formulated in different ways in a multimodal system, we can get completely different sets of retrieval results; hence needing the right combination method. Issues that are worth considering: how to intelligently combine the text and image queries to form hybrid queries; how to optimally combine retrieval result lists for multiple example query images; and how the text and image modalities can be optimally combined for a task without redundancy.
To address a few of the issues described above, and motivated by the successful use of machine learning and fusion techniques in IR, we propose a multimodal retrieval approach of biomedical images from collections of full-text journal articles. The proposed approach uses text and image features extracted from relevant components in a document, and exploits the hierarchical organization of the images based on classification and employs both intra- (visual feature level) and inter- (visual and textual result level) modality fusion techniques. The major contributions described in this article are:
-
A novel multimodal hierarchical modality classification approach for image filtering is proposed. The modality-specific information available from the classifier prediction on the query and database images is used to select the relevancy image subset.
-
A category-specific similarity fusion approach is used at image feature level. Individual pre-computed weights of different features are adjusted online based on the prediction of a query image modality, for a linear combination of similarity matching scheme.
-
A proposed fusion technique that takes into consideration multiple example query images to search for a particular information need.
-
A proposed inter-modality fusion of image and text-based result lists that considers past individual retrieval performance effectiveness.
-
And finally, a performed systematic retrieval evaluation in a standard benchmark ImageCLEFmed 2012 evaluation [12] dataset of more than 300,000 images with associated annotations that demonstrated significant improvement in performance comparatively.
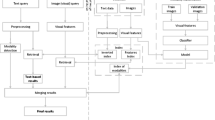
The block diagram of the proposed multi-modal retrieval process is shown in Fig. 2. Here, the top portion of the figure shows that a search is being initiated simultaneously based on both text and multiple query image parts of a multi-modal query/topic. In the middle portion of Fig. 2, it shows how different visual and textual features are extracted and presented for image classification and similarity matching. Finally, the bottom portion shows how individual image and text result lists are fused to obtain a final ranked list of top-matched images. Each of the sub-processes will be described more elaborately in the following sections.
The rest of the paper is organized as follows: in Sect. 2 we briefly describe the related works and their shortcomings in medical image retrieval. Textual and visual image feature extraction and representation is discussed in Sect. 3 and our image classification (modality detection) approach is discussed in Sect. 4. The content and text-based image retrieval approaches are described in Sects. 5 and 6, respectively. In Sect. 7, we describe the multi-modal search approach based on merging of visual and text result lists. The experiments and analysis of the results are presented in Sect. 8 and finally Sect. 9 provides the conclusions.
2 Related works
Conventional approaches to biomedical journal article retrieval have been text-based with little research done using images to improve text retrieval. For example, most retrieval tasks in biomedical literature use abstracts from MEDLINE [7]. On the other hand, retrieval systems searching for images within a collection of biomedical articles commonly represent and retrieve them according to their collateral text, such as captions [8, 9]. For example, the BioText [8] search engine; searches over 300 open access journals and retrieves figures as well as text. BioText uses the LuceneFootnote 6 text search engine to search full-text or abstracts of journal articles, as well as image and table captions. YottalookFootnote 7 allows multilingual searching to retrieve information (text or medical images) from the Web and journal articles. The goal of the search engine is to provide information to clinicians at the point of care. Other related work includes the Goldminer search engine that retrieves images by searching figure captions in peer-reviewed journal articles appearing in the Radiological Society of North America (RSNA) journals Radiographics and Radiology. It maps keywords in figure captions to concepts from the Unified Medical Language System (UMLS)Footnote 8 Metathesaurus. The Yale Image Finder (YIF) [9] searches text within images, captions, abstracts, and titles to retrieve images from biomedical journal papers. YIF uses optical character recognition (OCR) to recognize text in images in both landscape and portrait modes.
Most of the above systems do not use image features to find similar images or combine visual and text features for biomedical information retrieval. The IRMA system,Footnote 9 developed at Aachen University of Technology, Germany, aims to integrate text and image-based features for medical image retrieval. The system primarily uses visual features, but only uses a limited number of text labels that describe the anatomy, biosystem, image direction, and modality of the image. On the other hand, CBIR, for biomedical uses, has been studied extensively in academia and at research centers. The efforts focus on identifying subtle differences between images in homogenous collections that are often acquired as a part of health surveys or longitudinal clinical studies. For instance, the ASSERT system [18] is designed for high-resolution computed tomography (HRCT) images of the lung and the SPIRS system [19] for digitized X-rays of the spine.
The importance of medical illustrations in clinical decision-making has motivated the development of large databases of medical images, such as the Public Health Image Library (PHIL) and GoldMiner, as well as active research in image retrieval within the yearly ImageCLEF medical image retrieval tasks [10–12] and by individual researchers. There is also increasing commercial interest in multimodal information retrieval of biomedical articles as evidenced from the teams participating in the ImageCLEFmed contests. During the past several years, we witnessed several approaches for information fusion, especially text and visual search integration that have been used in ImageCLEF [10–12]. Progress in CBIR and retrieval based on text in image captions has motivated our research into integration of image data for semantic image retrieval.
3 Image representation
The performance of a classification and/or retrieval system depends on the underlying image representation, usually in the form of a feature vector. We represent each image as a combination of the textual and visual features described below:
3.1 Textual feature extraction and representation
We represent each image in the collection as a structured document of image-related text called an enriched citation. To generate structured documents required by our text search engine Essie [20], we augment MEDLINE citations with image-related information extracted from the full text, creating the enriched citations. Figure 3 depicts the enriched MEDLINE citation (PMID 18487544) of a short paper that contains no abstract and only one image (as shown in the left portion of the figure and referred in the \(<\)image\(>\) tag).
Our representation includes the title, abstract, and MeSH terms of the article in which the image appears as well as the image’s caption and mentions (snippets of text within the body of an article that discuss an image). Caption, abstract, and title extraction use document structure; rule-based methods are used for caption segmentation and mention extraction. The above-mentioned structured documents may be indexed and searched with a traditional search engine, or the underlying term vectors may be exposed and added to a mixed image representation for classification that includes the visual features as described in next section.
3.2 Visual feature extraction and representation
For content-based feature extraction, 14 different features were extracted to represent the images as shown in Table 1.
To represent the spatial structure of images, we utilized the Color Layout Descriptor (CLD) of MPEG-7 [22]. To represent the global shape/edge feature, the spatial distribution of edges were determined using the Edge Histogram Descriptor (EHD) [22]. The first (mean), second (standard deviation) and third (skewness) central moments of pixel intensities of gray-level images were calculated as color moments. Gabor descriptor is based on a multi-resolution decomposition using Gabor wavelets [24]. Tamura feature describes the coarseness, contrast, and directionality of an image and higher order moment-based texture feature was also extracted from the gray-level co-occurrence matrix (GLCM). A descriptor based on Scale-Invariant Feature Transform (SIFT) was computed from the intensity pattern within the affine covariant region [23]. Autocorrelation measures the coarseness of an image by evaluating the linear spatial relationships between texture primitives. The Lucene image retrieval (LIRE) library [26] is utilized to extract the Gabor, Tamura, Color edge directional descriptor (CEDD), and Fuzzy Color Texture Histogram (FCTH) features. In addition, two versions of Local Binary Pattern (LBP)-based feature are extracted [25]. The original LBP operator labels the pixels of an image by thresholding the 3-by-3 neighborhood of each pixel with the center pixel value and considers the result as a binary number. The 256-bin histogram of the labels computed over an image can be used as a texture descriptor. Each bin of histogram (LBP code) can be regarded as a micro-texton.
4 Image classification and modality detection
Image modality classification (detection) is an important task toward achieving high performance in biomedical image and article retrieval. Successful detection could play an important role in achieving better retrieval performance by reducing the search space to the set of relevant modalities. It can also enable the system to apply modality-specific algorithms to extract more useful and accurate information from the images for indexing and retrieval.
We implemented both flat and hierarchical classification, each of which uses textural, visual, or multimodal features. In the following, we describe our flat classification strategy, an extension of this approach that exploits the hierarchical structure of the classes, and a post-processing method for improving the classification accuracy of illustrations [29].
4.1 Flat classification
Owing to their empirical success, we utilized multi-class SVMs [30] for classifying images into 31 medical image modalities based on their textual and visual features. We composed multi-class SVMs using a one-against-one strategy. Among the many classification algorithms provided in WEKA, we found that Support Vector Machine (SVM)-based classification outperformed other classification algorithms for our classification purposes. For our SVM training, the input was a feature vector set of training images in which each image was manually annotated with a single modality label selected out of the \(M\) modalities. So, a set of \(M\) labels were defined as \(\{\omega _1,\ldots , \omega _i, \ldots , \omega _M\}\), where each \(\omega _i\) characterized the representative image modality.
First, we extracted our visual and textual image features from the training images (representing the textual features as term vectors). Then, we performed attribute selection to reduce the dimensionality of the features. We constructed the lower- dimensional vectors independently for each feature type (textual or visual) and combined the resulting attributes into a single, compound vector. The dimensions (number of attributes) of visual and textual features before attribute selection were 1,361 and 2,703, respectively. Finally, we used the lower-dimensional feature vectors to train multi-class SVMs for producing textual, visual, or mixed modality predictions.
4.2 Hierarchical classification
Unlike the flat classification strategy described above, it is possible to exploit the hierarchical organization of the modality classes in order to decompose the task into several smaller classification problems that can be sequentially applied. Based on our visual observation of the training samples and on our initial experiments, we modified the original modality hierarchy (as shown in Fig. 4 and as reported in Müller et al. [27]) for ImageCLEFmed 2012 classification task [12] to a new hierarchy with the same acronyms for classes as shown in Fig. 5.
Modality classes in the biomedical literature [27]
Our proposed hierarchy for hierarchical classification (acronyms defined in Fig. 4)
We implemented a classifier that classifies images into one of the 30 modality classes (excluding COMP). This flat 30-class classifier was sufficient to identify modality classes that were frequently misclassified by our visual features. For example, samples in “Non-clinical photos” (GNCP) under the “Generic biomedical illustrations” in Fig. 4 were basically photos and they were frequently misclassified into one of the three modality classes in “Visible light photography” under “Diagnostic images”. We moved GNCP to the “Photo” category in our hierarchy to prevent classification errors at the upper level. For the same reason, three modality classes under “Printed signals, waves” were moved to “Illustration”, and “3D reconstructions” was merged into the Radiology category.
In our proposed hierarchy, we first separated illustration modality classes from others. Compound images (COMP) were then considered separately within the two categories, “Illustration” and “General”; however, they needed to be merged into one list, i.e., COMP modality class. Under “Illustration/Single” all 14 modality classes were classified by one flat classifier and no more lower levels existed. “General/Single” category has three sub-categories, viz., “Radiology_3D”, “Photo”, and “Microscopic”, where samples in each category were acquired by identical or similar imaging technology or had similar visual features. Each category under the “General/Single” was then expanded into their leaf modality classes which were the final modality classes in the task.
Six classifiers were implemented for the classification task in each level. We trained flat multi-class SVMs for each meta-class. We combined all 15 visual features and then selected the most relevant attributes for each classification task by an attribute selection function in WEKA [31]. Hence, every classifier used a different set of attributes. A tenfold cross-validation (CV) was selected for evaluation since several modality classes in the training set had insufficient samples (number of samples in each class ranged from 5 to 49).
For recognizing compound images (e.g., “COMP” class), we utilized the algorithm proposed in [28], which detects sub-figure labels (e.g., ‘A’, ‘B’, ‘C’, etc.), if present, and the border of each sub-figure within a compound image. To arrive at a final class label, an image was sequentially classified beginning at the root of the hierarchy until a leaf class was determined. An advantage of performing hierarchical classification was that we could filter the retrieved results using the meta-classes within the hierarchy (e.g., “Radiology”).
4.2.1 Illustration post-processing
Because our initial classification experiments resulted in only modest accuracy for the 14 “Illustration” classes shown in Fig. 5, we concluded that our textual and visual features may not have been sufficient for representing these figures. Therefore, in addition to the aforementioned machine learning modality classification methods, we also developed several complimentary rule-based strategies for increasing the classification accuracy of “Illustration” classes. The majority of the training samples contained in the “Illustration” meta-class, unlike other images in the collection, consisted of line drawings or text superimposed on a white background. For example, program listings mostly consisted of text; thus, we anticipated that the use of text and line detection methods could increase the classification accuracy of Class “GPLI”. Similarly, polygons (e.g., rectangles, hexagons, etc.) contained in flowcharts (GFLO), tables (GTAB), system overviews (GSYS), and chemical structures (GCHE) were a distinctive feature of these modalities. We utilized the methods of Jung et al. [32] and OpenCVFootnote 10 functions to assess the presence of text and polygons, respectively.
5 Content-based image retrieval (CBIR) approach
Our content-based approach to image retrieval was based on retrieving images that appeared visually similar to the given topic images. In CBIR, access to information was performed at a perceptual level based on automatically extracted low-level features (e.g., color, texture, shape, etc.). The retrieval performance of CBIR mainly depended on the underlying image representation, usually in the form of a feature vector described in Sect. 3.2. It was challenging to find a unique feature representation to compare images accurately for all types of queries. Feature descriptors at different levels of image representation were in diverse forms and usually complementary in nature.
5.1 Category-specific fusion
The CBIR community adopts some of the ideas of the data fusion research in document retrieval. The most commonly used approach is the linear combination of similarity matching of different features with pre-determined weights. In this framework, the similarity between a query image \(I_q\) and target image \(I_j\) is described as
where \(F \in \{\hbox {CLD, EHD, CEDD, etc.}\}\) and \({\hbox {S}}^{F}(I_q, I_j)\) are the similarity-matching function in individual feature spaces and \(\alpha ^{F}\) are weights (generally decided by users or hard coded in the systems) within the different image representation schemes (e.g., intra-modality weights). However, there is a limitation with the above hard-coded or fixed-weight-based similarity matching approach. In this approach, for example, a color feature has the same weight for a search of the microscopic pathology or X-ray images. Although color is an important feature for microscopic and photographic images, it is not a reliable feature for X-ray images.
To overcome this limitation, we explored a query-specific adaptive linear combination of similarity fusion approach by relying on the image classification information [13]. In this approach, for a query image, its category at a global level (e.g., modality) was predicted by using the classifiers discussed in Sect. 4. Based on the online category prediction of a query image, pre-computed category-specific feature weights (e.g., \(\alpha ^{F}\)) were utilized in the linear combination of the similarity-matching function. Based on this scheme, for example, a color feature had more weight for microscopic pathology and photographic images, whereas edge and texture-related features had more weight for radiographs and illustrations.
5.2 Image filtering
It is computationally expensive to perform a linear image search in a large collection. To overcome this, we utilized the modality information of query and database images for image filtering to reduce the search space. The similarity-matching approach described above was only performed if the modality of a query image matched with an image in the collection. During the feature extraction process of images in the collection, the image categories were determined by applying the SVM classification scheme based on the individual image feature input (described in Sect. 4) and the information stored along with the feature indices in a logical database. Similar feature extraction and category prediction stages were performed online when the search is performed with an unknown query image. The modality of the query image and the database images from the category index are quickly evaluated to identify candidate target images in the collection, thereby filtering out irrelevant images from further consideration.
For image filtering and fusion, the first two levels in Fig. 5 were considered as a trade-off between time and accuracy. The cross-validation accuracies were above or around 95 % for Classifier 1 and Classifier 2 as shown in Table 3 (and discussed in the Results section). If we had considered more deeper levels for modality categorization, then search might have been faster, but at the expense of lower classification accuracies at leaf levels and as a consequence of retrieval accuracies.
The steps involved in the above fusion and filtering processes are depicted in Algorithm 1.

From the \(IF\) step of the Algorithm 1, we observed that only pre-computed category-specific feature weights (e.g., \(\alpha ^{F}\)) were utilized for similarity matching with filtered database images when their modality matched to the modality of the query image.
6 Text-based image search approach
For the text-based image search, we indexed our enriched citations described in Sect. 3.1 with the Essie search engine [20] developed by the National Library of Medicine (NLM). It is particularly well-suited for the medical retrieval task due to its ability to automatically expand query terms using the UMLS synonymy and its ability to weight term occurrences according to the location in the document in which they occur. For example, term occurrences in an image caption can be given a higher weight than occurrences in the abstract of the article in which the image appears. Essie also expands query terms to include morphological variants derived from the UMLS SPECIALIST Lexicon instead of stemming. Essie’s algorithm for scoring the similarity between a document and a query can be summed up as preferring “all the right pieces in all the right places”. The “right pieces” are phrases from the query, and the “right places” are the fields of a document most valuable for a retrieval task, such as image captions for image retrieval, or MeSH for literature retrieval.
To construct a query for each topic, we created and combined several Boolean expressions derived from the extracted concepts. First, we created an expression by combining the concepts using the logical \(AND\) operator (i.e., all concepts were required to occur in an image’s textual representation), and then we produced additional expressions by allowing an increasing number of the extracted concepts to be optional. Finally, we combined these expressions using the logical \(OR\) operator giving significantly more weight to expressions containing a fewer number of optional concepts. Additionally, we included a verbatim topic description as a component of a query, when needed, but we gave minimal weight to this expression compared to those containing the extracted concepts. The resulting queries were then used to search the Essie indices.
7 Multimodal search based on merging
For the multimodal search, we directly combined the independent outputs of our textual and content-based search results. The “Dark Horse Effect” in data fusion [21] assumes that a good fusion algorithm should treat the systems which retrieve a larger number of relevant images differently than other systems which do not retrieve a large number of relevant images. This means that we should give more importance (or weight) to a retrieval system based on the number of relevant images it has retrieved. By observing the retrieval results of the ImageCLEF track during the past several years [10, 11], we concluded that the text-based retrieval systems overwhelmingly outperformed visual systems in terms of precision and other measures. It was therefore important to determine optimal fusion strategies, allowing overall performance improvement over the constituent system as in the past some groups had combinations leading to poorer results than using textual retrieval alone. The weights might have been set manually, adjusted on-line, or learned off-line according to this prior knowledge, which was clearly evident from many fusion methods that were presented in ImageCLEF track sessions.
For our multi-modal fusion, we used a linear combination method, where weights of the individual retrieval systems are determined by a function of their performance measured by the Mean Average Precision (MAP) values. In this regard, for off-line learning on training samples we used our best image and text-based MAP scores from the previous year (e.g., CLEF’2011). The raw MAP scores were normalized by the total score as \( \omega _I = \frac{\mathrm{MAP} (I)}{\mathrm{MAP} (I) + \mathrm{MAP} (D)} \) and \(\omega _D = \frac{\mathrm{MAP} (D)}{\mathrm{MAP} (I) + \mathrm{MAP} (D)} \) to generate the image and text feature weights, respectively.
For multi-modal retrieval purposes, we consider ed \(q\) as a composite query which has an image part as \(I_q\) and a text part \(D_q\) as enriched citation. In a linear combination scheme, the similarity between \(q\) and a multi-modal item \(j\), which also has two parts (e.g., image \(I_j\) and text \(D_j\)), was defined as
where \(\omega _I\) and \(\omega _D\) are inter-modality weights within the image and text feature spaces as described above, which are subject to \(0 \le \omega _I, \omega _D \le 1\) and \(\omega _I + \omega _D = 1\). In this framework, the image-based similarity \(\hbox {Sim} (I_q , I_j)\) was already defined in (1) and the text-based similarity \(\hbox {Sim}(D_q , D_j)\) score was based on the results returned by the Essie [20] search engine.
In the context of ImageCLEF evaluation, each ad hoc topic contained a short sentence or phrase describing the search request in a few words with one to several relevant sample images. For our multi-modal search approach, the description of the topics were used as the search terms to search NLM’s Essie search engine [20] and sample images were utilized as “Query By Example (QBE)” for the CBIR search. For the CBIR search of several sample query images of a topic, we obtained separate ranked result lists. In this scenario, to obtain a final ranked list of images based on purely visual search, we performed a CombMax fusion on similarity scores on the individual result lists by considering only the top 1,000 images. The reason for using CombMAX was that images which matched closely with at least one query image was ranked highly and was more effective when sample query images were diverse in modality in many semantic topic categories. As already mentioned, CombMAX favors images highly ranked in one system (e.g., the aforementioned Dark Horse Effect).
Finally, the image and text-based result lists were merged to produce a combined ranked result list by applying the weighted linear combination scheme based on normalized MAP scores. Since the content and text-based result lists were created from two completely different independent systems, their similarity scores were normalized before performing any fusion by applying a Min-Max normalization [16]. To consider the above effect and explore further, we also performed a rank to score based merging for our multi-modal search approach. In this approach, the rank of the first 1,000 images in the result lists were considered and converted to similarity score by using the formula \((1- (\mathrm{rank} (\mathrm{image})/1{,}000))\). Hence, an image with rank position 1 (e.g., rank (image) = 1) had a score of 0.999 and for the rank position 1,000, the score was 0.
The overall multimodal search approach from a topic perspective of ImageCLEF’2012 is described in steps in Algorithm 2.

8 Experiments and results
This section presents the detailed empirical analysis of the proposed classification and retrieval techniques described in the above sections. Specially, we present the data sets used for the experiments, experimental settings, accuracy comparisons of classification and retrieval, and analysis of the results.
8.1 Image collections
For the purposes of this research, we used the ImageCLEFmed 2012 dataset [12] which contains over 300,000 images from 75,000 biomedical journal articles in the open access literature subset of the PubMed CentralFootnote 11 database. The contents of this collection represent a broad and significant body of medical knowledge, which made the retrieval more challenging. The collection contains a variety of imaging modalities, image sizes, and resolutions and can be considered as a fairly realistic set for evaluating medical image retrieval techniques. The experimental results were generated based on 22 ad hoc topics divided into visual, mixed and semantic types. Each topic consisted of the query itself in three languages (English, German, French) and one to few sample images for the visual part of the topic. The topics were based on a selection of queries from search logs of the Goldminer radiology image search system [33].
Figure 6 shows an example of a topic description “renal vein thrombosis” along with a few sample images from ImageCLEFmed 2012 evaluation [12]. We could easily observe the large variation in visual appearances of the images of different modality types, which made the content-based visual search difficult.
A training set of 1,000 images from 31 categories was provided by the ImageCLEFmed 2012 organizers for classifier training. On average, there were over 30 images per category, although a few categories (such as; “DSEC”, “DSEE”, “DSEM”) had \(<\)10 images, which made the training set non-uniform in nature and much harder for the learning algorithm.
8.2 Performance measures
The relevant sets of all topics were created by the CLEF organizers by considering the top retrieval results of all submitted runs of the participating groups. Results for submitted runs were computed using the latest version of TREC-EVALFootnote 12 software. Results were evaluated using un-interpolated (arithmetic) mean average precisions (MAP) to test effectiveness and precisions at different ranks (e.g., P5, P10, P20, etc.). Further measures considered included geometric mean average precision (GMAP) to test robustness, and the Binary Preference (B-PREF) measure which is a good indicator for the completeness of relevance judgments. The performances were compared for different feature spaces (e.g., textual and visual) and with and without using image filtering.
8.3 Classification result
The results of the modality classification approaches were compared using classification accuracy. Table 2 shows our overall modality classification results based on 1,000 test images provided by the CLEF organizers. We submitted nine different runs, out of which six representative runs are shown here. Hierarchical classification showed slightly better performance than flat classification and using multi-modal features, achieved about 10–20 % higher accuracy than individual features. We achieved the highest accuracy (63.2 %) for our submitted runs by applying multi-modal, hierarchical, and post-processing methods as described in Sect. 4. Our best run ranked within the submissions from the top three participating groups. This result validates our post-processing method used to improve the recognition of “Illustration” classes, and provides, with our previous experience [17], further evidence that hierarchical classification is a successful strategy. Each of our hierarchical classification methods outperformed the corresponding approach having the same feature representation. The results also show that the visual features outperform the textual features when they are used individually.
While our submitted runs were only judged on their ability to identify each of the 31 modality classes [27], Table 3 presents the classification accuracy of the intermediate classifiers we used for our hierarchical approaches. Table 3 gives the number of classes contained for each meta-class in the hierarchy shown in Fig. 5 and the classification accuracy associated with the textual, visual, and mixed feature representations.
As shown in the Table 3, when multi-modal features were used, Classifier 1 and 2 achieved over 90 % accuracy and Classifier 3, 4, and 5 achieved more or \(<\)80 % accuracy. However, Classifier 6 (Illustration classifier) achieved the lowest accuracy of 63.49 %. The post-processing method for “Illustration” increased the accuracy for Classifier 6 and for the entire classification at about 7 and 3 %, respectively. In each classifier, classification errors from higher classifiers in the hierarchy are accumulated.
These results also demonstrate that the accuracies of the intermediate classifiers generally improved as the number of class labels decreased. Given the limited amount of training data in relation to the number of total modalities, the smaller number of labels per classifier likely is significant for explaining why our hierarchical classification approaches consistently outperformed their corresponding approaches.
8.4 Retrieval result
Table 4 shows the various performance measures of different search schemes as proposed. It is clear from Table 4 that the best MAP score (0.2533) was achieved when a multi-modal search (weighted score-based) was performed in a filtered image set. When the multi-modal search approach was compared with individual text and visual-based approaches, we observed that almost every score (e.g., MAP, GM-MAP, Rprec, etc.) improved by considering the counterparts. Overall, the performance of the CBIR search approach was very low compared to the text-based and multi-modal searches as observed in Table 4. The main reason was the high-level semantic contents of query topics. This result might be an indication that the query topics are more semantic in nature and this is always the case for the ImageCLEF evaluation campaign. However, it is evident from the results of the Table 4 that combining both the visual and text-based searches can improve retrieval performance.
To breakdown the results further, Fig. 7 shows the bar graph (chart) of different search approaches based on precision over the top K (5, 10, and 20) images. We observed significant improvement of precision at these early ranks for the multi-modal search approach when compared to individual visual and text-based search approaches. It strongly validates the point that for semantic retrieval of images in the medical domain, associated or contextual information largely improves retrieval precision.
Figure 8 shows the bar graphs of MAP scores for CBIR and multi-modal (weighted score and ran-score based) searches with and without using image filtering. Although the MAP score was slightly decreased for the CBIR search with filtering, it was slightly increased for both the multi-modal searches as shown in the figure. Finally, from the results, we can also conjecture that the pre-filtering approach is indeed an effective one for our multi-modal search approach when compared to the search which was performed on the entire collection.
Further, an important benefit of searching on a filtered image set is gain in computation time. We tested the efficiency of the multi-modal search scheme by comparing the average retrieval time for 22 query topics with and without applying the filtering scheme. The experiment was performed in an Intel Pentium Dual-Core CPU at 3.40 GHz with 12 GB of RAM running Microsoft Windows 7 operating system. The linear search time without filtering was twice as much as search on the filtered image set, suggesting that the proposed filtering scheme is both effective and efficient. The average retrieval time for the topics is currently 1.3 s, which includes the computational cost for feature extraction, classification, similarity matching for CBIR and text retrieval, and merging of the result lists. Without filtering the computational cost can further increase as the database grows due to our linear search scheme for content-based similarity matching. However, by using some multi dimensional indexing scheme and parallelizing the feature extraction processes, we plan to reduce the search time in future.
In some cases, combining image and text-based search results might also have negative effects in the final retrieval result. For example, the topic in Fig. 6 is a “semantic” type with perceptually very different sample images based on modality and appearances. This topic should be well suited for textual retrieval approach only, and integrating or refining it with a content-based search would only decrease the performance of a final retrieval result set. Our multi-modal search approaches are more suited and tailored for mixed-mode topics where both visual and text-based search can contribute to the final result.
To prove this argument, our search results were also tested and compared by considering only 14 mixed type topics to the 22 topics for ImageCLEFmed 2012 evaluation. Figures 9 and 10 show the bar graphs of MAP and P5 scores for searches in different feature spaces (e.g., visual, text, and multimodal) for “Mixed” only and “All” query topics. We observed a significant improvement in MAP and P5 scores for “Mixed” topics for both multi-modal and text-based search approaches, and a decrease in P5 score for CBIR for “Mixed” topics when compared to the search on “All” topics. Basically it shows that “Mixed” type queries are more suitable for text or multi-modal search approaches and CBIR is only good when the topic is only “Visual” type.
Finally, a comparison of results from the top five multi-modal approaches by other ImageCLEFmed’2012 participants is shown in Table 5. The best run here with a MAP score of 0.2377 was submitted by our group [34]. This multi-modal approach uses a textual representation of visual features (image cluster words) described in Sect. 3.2 that was easily integrated with our existing textual features. Comparing these top runs with our best proposed run with a MAP score of 0.2533 as shown in Table 4, using Fisher’s paired randomization test [35], a recommended statistical test for evaluating information retrieval systems, it was found that we achieved a statistically significant increase (6.5 %, \(p = 0.024330\)) over the top-ranked performance in ImageCLEFmed 2012.
9 Conclusions
Information extraction and retrieval are essential tasks required for achieving many of the ultimate goals of biomedical informatics research and development. In this paper, a novel multi-modal retrieval approach for biomedical articles is proposed inspired by the ideas of IR, CBIR, NLP, and Machine Learning paradigms. Unlike many other approaches, where the search is performed with a single modality and without any classification information, we propose to use the classification result directly in the retrieval loop and fuse the results effectively obtained from both the text and imaging modalities. In particular, we present ways to improve retrieval performance by making use of textual as well as visual information. A standard data set with a query set and corresponding performance measure model, such as the ImageCLEFmed 2012 collection has provided enough reliability for objective performance evaluation. Our retrieval results demonstrate the effectiveness and efficiency of the proposed multi-modal framework compared to using only a single modality or without using any classification information. Due to the multi-disciplinary and multi-perspective nature of this work, we have a good opportunity to expand our work. In the future, we want to focus on constructing a model or formalism to show how the inclusion of text can contribute to the improvement of image retrieval or vice versa. A major issue is scalability and efficiency. Since we use a large image collection and several query and image representations for different types of fusion, a large computational overhead currently persists. To overcome this, we will concentrate more on a multi-dimensional and especially multi-feature indexing approach, which might provide a challenging topic for our retrieval research.
Notes
MeSH is a controlled vocabulary created by the U.S. National Library of Medicine to index biomedical articles.
References
Demner-Fushman D, Antani SK, Simpson M, Thoma GR (2009) Annotation and retrieval of clinically relevant images. Int J Med Inf 78(12):e59–e67
Demner-Fushman D, Antani SK, Simpson M, Thoma GR (2012) Design and development of a multimodal biomedical information retrieval system. JCSE 6(2):168–177
Antin-Ozerkis D, Rubinowitz A (2008) Recognizing lung disease in patients with rheumatoid arthritis. Part 2: pleuropulmonary disease may be more common than you thought. J Respir Dis 29(8):318–324
Datta R, Joshi D, Li J, Wang JZ (2008) Image retrieval: ideas, influences, and trends of the new age. ACM Comput Surv 40(2): 1–60
Chen N (2006) A survey of indexing and retrieval of multimodal documents: text and images. Technical Report, 2006–505, Queen’s University
Ingwersen P (1996) Cognitive perspectives of information retrieval interaction: elements of a cognitive IR theory. J Doc 52(1): 3–50
Hersh WR et al (2004) TREC 2004 genomics track overview. The Thirteenth Text Retrieval Conference: TREC 2004. National Institute of Standards and Technology, Gaithersburg
Hearst MA, Divoli A, Buturu H, Ksikes A, Nakov P, Wooldridge MA et al (2007) Biotext search engine: beyond abstract search. Bioinformatics 23(16):2196–2197
Xu S, McCusker J, Krauthammer M (2008) Yale Image Finder (YIF): a new search engine for retrieving biomedical images. Bioinformatics 24(17):1968–1970
Müller H, Kalpathy-Cramer J, Eggel I, Bedrick S, Reisetter Jr J, Khan CEKJ, Hersh WR (2010) Overview of the CLEF 2010 Medical Image Retrieval Track. In: CLEF 2012 Evaluation Labs and Workshop, Online Working Notes, Padua, Italy, 20–23 Sept 2010
Kalpathy-Cramer J, Müller H, Bedrick S, Eggel I, Garcia Seco de Herrera A, Tsikrika T (2011) Overview of the CLEF 2011 medical image classification and retrieval tasks. In: CLEF 2011 Working Notes, Amsterdam, The Netherlands
Müller H, Herrera A, Kalpathy-Cramer J, Demner-Fushman D, Antani S, Ivan E (2012) Overview of the ImageCLEF 2012 Medical Image Retrieval and Classification Tasks. In: The Working Notes for the CLEF 2012 Labs and Workshop, Rome, Italy, 17–20 Sept 2012
Rahman MM, Antani SK, Thoma GR (2011) A learning-based similarity fusion and filtering approach for biomedical image retrieval using SVM classification and relevance feedback. IEEE Trans Inf Technol Biomed 15(4):640–646
Lehmann TM, Güld MO, Deselaers T, Keysers D, Schubert H, Spitzer K, Ney H, Wein BB (2005) Automatic categorization of medical images for content-based retrieval and data mining. Comput Med Imaging Graph 29:143–155
Fox EA, Shaw JA (1994) Combination of multiple searches. In: Proceedings of the 2nd Text Retrieval Conference (TREC-2), NIST Special Publication, pp 243–252
Lee JH (1995) Combining multiple evidence from different properties of weighting schemes. In: Proceedings of the 18th Annual ACM-SIGIR, pp 180–188
Simpson M, Rahman MM, Phadmis S, Apostolova E, Demner-Fushman D, Antani SK, Thoma GR (2011) Text- and content-based approaches to image modality classification and retrieval for the ImageCLEF 2011 medical retrieval track. In: The Working Notes for the CLEF 2011 Labs and Workshop, Amsterdam, 19–22 Sept 2011
Shyu CR, Brodley CE, Kak AC, Kosaka A, Aisen AM, Broderick LS (1999) ASSERT: a physician-in-the-loop content-based image retrieval system for HRCT image databases. Comput Vis Image Underst 75:111–132
Hsu W, Antani S, Long LR, Neve L, Thoma GR (2008) SPIRS: a web-based image retrieval system for large biomedical databases. Int J Med Inf 78:13–24
Ide NC, Loane RF, Demner-Fushman D (2007) Essie: a concept-based search engine for structured biomedical text. J Am Med Inf Assoc 1(3):253–263
Baeza-Yates R, Ribeiro-Neto B (June 1999) Modern information retrieval. Addison-Wesley, Reading
Chang SF, Sikora T, Puri A (2001) Overview of the MPEG-7 standard. IEEE Trans Circ Syst Video Technol 11:688–695
Lowe DG (2004) Distinctive image features from scale-invariant keypoints. Int J Comput Vis 60(2):91–110
Manjunath BS, Ma WY (1996) Texture features for browsing and retrieval of large image data. IEEE Trans Pattern Anal Mach Intell (Special Issue on Digital Libraries) 18(8):837–842
Menp T (2003) The local binary pattern approach to texture analysis extensions and applications. PhD thesis, University of Oulu
Grubinger M, Clough P, Hanbury A, Müller H (2008) Lire: lucene image retrieval: an extensible java CBIR library. In: Proceedings of the 16th ACM international conference on Multimedia, Vancouver, British Columbia, Canada, pp 1085–1088
Müller H, Kalpathy-Cramer J, Demner-Fushman D, Antani S (2012) Creating a classification of image types in the medical literature for visual categorization. In: Advanced PACS-based Imaging Informatics and Therapeutic Applications, Proceedings of the SPIE, vol 8319, pp 83190P–83190P-12
Apostolova E, You D, Xue Z, Antani S, Demner-Fushman D, Thoma GR (2013) Image retrieval from scientific publications: text and image content processing to separate multi-panel figures. J Am Soc Inf Sci Technol. Published online in Wiley Online Library (wileyonlinelibrary.com). doi:10.1002/asi.2281
You D, Rahman MM, Antani SK, Demner-Fushman D, Thoma GR (2013) Text- and content-based biomedical image modality classification. In: SPIE Proceedings vol 8674 Medical Imaging, Advanced PACS-based Imaging Informatics and Therapeutic Applications. Maria Y. Law; William W, Boonn, Editors 86740L
Vapnik V (1998) Statistical learning theory. Wiley, New York
Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH (2009) The WEKA data mining software: an update. SIGKDD Explor 11(1):10–18
Jung K, Kim KI, Jain AK (2004) Text information extraction in images and video: a survey. Pattern Recognit 37(5):977–997
Tsikrika T, Müller H, Kahn Jr CE (2012) Log analysis to understand medical professionals’ image searching behaviour. In: Proceedings of the 24th European Medical Informatics Conference (MIE2012)
Simpson M, You D, Rahman MdMM, Demner-Fushman D, Antani Sk, Thoma GR (2012) ITI’s participation in the ImageCLEF 2012 medical retrieval and classification tasks. In: The Working Notes for the CLEF 2012 Labs and Workshop, Amsterdam, Rome, Italy, 17–20 Sept 2012
Smucker MD, Allan J, Carterette B (2007) A comparison of statistical significance tests for information retrieval evaluation. In: Proceedings of the Sixteenth ACM Conference on Information and Knowledge Management, pp 623–632
Acknowledgments
This research is supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM), and Lister Hill National Center for Biomedical Communications (LHNCBC).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rahman, M.M., You, D., Simpson, M.S. et al. Multimodal biomedical image retrieval using hierarchical classification and modality fusion. Int J Multimed Info Retr 2, 159–173 (2013). https://doi.org/10.1007/s13735-013-0038-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13735-013-0038-4