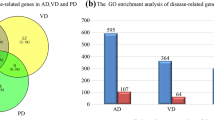
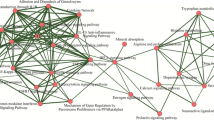
Abstract
In the world, Huntington’s disease, Parkinson’s disease, and Alzheimer’s disease are reported as the most deadly diseases to issue common disorders for human beings. In state-of-the-art works, it is well studied that Huntington’s disease (HD) and Parkinson’s disease (PD) are affected by genetic factors and common motor system disorder, respectively. On the other hand, Alzheimer’s disease (AD) is a neurodegenerative disorder which is categorized by age-related dementia, behavioral changes, and memory loss, etc. In the literature, it is also identified that lots of similar genetic factors are interrelated among these diseases. In this paper, our key objective is to screen out the most significant common gene targets among HD, PD, and AD diseases. Here, the analysis attempts to discover efficient drug design for reactive genes by analyzing the topological properties of protein–protein interaction (PPI) networks. Initially, the analysis is started by reducing the number of total reactive genes (around 37 common genes) through preprocessing and cross-linkage. Afterward, we have identified five most significant hub proteins among these three diseases based on degree value. Moreover, a gene regulatory network has been designed to explain molecular mechanisms that significantly help to discover drugs for targeted genes.








(available at https://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index) and themiRTarBase (available at ) database

(available at https://www.encodeproject.org/) database

(available at https://regnetworkweb.org/)

(available at https://www.drugbank.ca/) database

(available at https://www.drugbank.ca/) database

(available at https://www.drugbank.ca/) database

(available at https://ctdbase.org/)
Similar content being viewed by others
References
Ahmed ML, Islam MR, Paul BK, Ahmed K, Bhuyian T (2020) Computational modeling and analysis of gene regulatory interaction network for metabolic disorder: a bioinformatics approach. Biointerface Res Appl Chem 10(4):5910–5917
Bolgár B, Antal P (2017) VB-MK-LMF: fusion of drugs, targets and interactions using variational Bayesian multiple kernel logistic matrix factorization. BMC Bioinform 18(1):440
Burns A, Iliffe S (2009) Alzheimer’s disease. BMJ 338(7692):467–471
Caufield JH, Wimble C, Shary S, Wuchty S, Uetz P (2017) Bacterial protein meta-interactomes predict cross-species interactions and protein function. BMC Bioinform 18(1):171
Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK (2008) Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimer’s Dis 14(1):27–41
Cohen B, Oren M, Min H, Perl Y, Halper M (2008) Automated comparative auditing of NCIT genomic roles using NCBI. J Biomed Inform 41(6):904–913
Dayalu P, Albin RL (2015) Huntington disease: pathogenesis and treatment. Neurol Clin 33(1):101–114
De Lau LM, Breteler MM (2006) Epidemiology of Parkinson’s disease. Lancet Neurol 5(6):525–535
Friedman N (2004) Inferring cellular networks using probabilistic graphical models. Science 303(5659):799–805
Hasan MR, Paul BK, Ahmed K, Mahmud S, Dutta M, Hosen MS, Hassan MM, Bhuyian T (2020) Computational analysis of network model based relationship of mental disorder with depression. Biointerface Res Appl Chem 10(5):6293–6305
Hughes AJ, Daniel SE, Blankson S, Lees AJ (1993) A clinicopathologic study of 100 cases of Parkinson’s disease. Arch Neurol 50(2):140–148
Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N (2002) Revealing modular organization in the yeast transcriptional network. Nat Genet 31(4):370–377
Lang AE, Lozano AM (1998) Parkinson’s disease. N Engl J Med 339(16):1130–1143
Lomax J, McCray AT (2004) Mapping the gene ontology into the unified medical language system. Int J Genom 5(4):354–361
Malkhasian AY, Howlin BJ (2019) Automated drug design of kinase inhibitors to treat Chronic Myeloid Leukemia. J Mol Graph Model 91:52–60
Manoharan S, Guillemin GJ, Abiramasundari RS, Essa MM, Akbar M, Akbar MD (2016) The role of reactive oxygen species in the pathogenesis of Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease: a mini review. Oxid Med Cell Longev.
Mantle D, Falkous G, Ishiura S, Blanchard PJ, Perry EK (1996) cc. Clinicachimicaacta 249(1–2):129–139
Mendez MF (2012) Early-onset Alzheimer’s disease: nonamnestic subtypes and type 2 AD. Arch Med Res 43(8):677–685
Morelli X, Bourgeas R, Roche P (2011) Chemical and structural lessons from recent successes in protein–protein interaction inhibition (2P2I). Curr Opin Chem Biol 15(4):475–481
Rajput AH, Rozdilsky BL, Rajput A (1991) Accuracy of clinical diagnosis in parkinsonism—a prospective study. Can J Neurol Sci 18(3):275–278
Ray S, Hossain SMM, Khatun L, Mukhopadhyay A (2017) A comprehensive analysis on preservation patterns of gene co-expression networks during Alzheimer’s disease progression. BMC Bioinform 18(1):579
Roos RA, Hermans J, Vegter-Van Der Vlis M, Van Ommen GJ, Bruyn GW (1993) Duration of illness in Huntington’s disease is not related to age at onset. J Neurol Neurosurg Psychiatr 56(1):98–100
Sachs K (2005) Causal protein-signaling networks derived from multiparameter single-cell data. Science 308(5721):523–529. https://doi.org/10.1126/science.1105809
Satagopan JM, Venkatraman ES, Begg CB (2004) Two-stage designs for gene–disease association studies with sample size constraints. Biometrics 60(3):589–597
Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N (2003) Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 34(2):166–176
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Sperandio O, Reynès CH, Camproux AC, Villoutreix BO (2010) Rationalizing the chemical space of protein–protein interaction inhibitors. Drug Discov Today 15(5–6):220–229
Walker FO (2007) Huntington’s disease. Lancet 369(9557):218–228
Wang X, Ren X, Li B, Yue J, Liang L (2012) Applying modularity analysis of PPI networks to sequenced organisms. Virulence 3(5):459–463
Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38(2):W214–W220
Wilson RS, Barral S, Lee JH, Leurgans SE, Foroud TM, Sweet RA, Graff-Radford N, Bird TD, Mayeux R, Bennett DA (2011) Heritability of different forms of memory in the late onset Alzheimer’s disease family study. J Alzheimer’s Dis 23(2):249–255
Xia J, Gill EE, Hancock RE (2015) NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protocols 10(6):823
Young TE (2012) Therapeutic drug monitoring–the appropriate use of drug level measurement in the care of the neonate. Clin Perinatol 39(1):25–31
Yu D, Lim J, Wang X, Liang F, Xiao G (2017) Enhanced construction of gene regulatory networks using hub gene information. BMC Bioinform 18(1):186
Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genetics Mol Biol, 4(1).
Zhu M, Gao L, Li X, Liu Z, Xu C, Yan Y, Walker E, Jiang W, Su B, Chen X, Lin H (2009) The analysis of the drug–targets based on the topological properties in the human protein–protein interaction network. J Drug Target 17(7):524–532
Acknowledgements
This manuscript has not been published yet and not even under consideration for publication elsewhere. The authors are grateful who have participated in this research work.
Funding
There is no funding for this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors have read the manuscript and approved this for submission as well as no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kawsar, M., Taz, T.A., Paul, B.K. et al. Identification of vital regulatory genes with network pathways among Huntington’s, Parkinson’s, and Alzheimer’s diseases. Netw Model Anal Health Inform Bioinforma 9, 50 (2020). https://doi.org/10.1007/s13721-020-00257-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-020-00257-4