Abstract
Prediction is one of the most attractive aspects in data mining. Link prediction has recently attracted the attention of many researchers as an effective technique to be used in graph based models in general and in particular for social network analysis due to the recent popularity of the field. Link prediction helps to understand associations between nodes in social communities. Existing link prediction-related approaches described in the literature are limited to predict links that are anticipated to exist in the future. To the best of our knowledge, none of the previous works in this area has explored the prediction of links that could disappear in the future. We argue that the latter set of links are important to know about; they are at least equally important as and do complement the positive link prediction process in order to plan better for the future. In this paper, we propose a link prediction model which is capable of predicting both links that might exist and links that may disappear in the future. The model has been successfully applied in two different though very related domains, namely health care and gene expression networks. The former application concentrates on physicians and their interactions while the second application covers genes and their interactions. We have tested our model using different classifiers and the reported results are encouraging. Finally, we compare our approach with the internal links approach and we reached the conclusion that our approach performs very well in both bipartite and non-bipartite graphs.
Similar content being viewed by others
1 Introduction
A network is characterized by a set of nodes and the links connecting them. Links are determined based on the scope covered by the network. Networks exist intensively in a wide range of domains from computing to systems biology to sociology, etc. Social network modeling is important for understanding organizational structures, group behavior, and the nature of social ties in propagating information. Amongst a variety of application domains, link prediction is one of the most challenging issues in social network research. The majority of link prediction methods focus on the emergence of links or social ties originating from the network structure, i.e., modules or hierarchies. In this paper, we think in a more comprehensive way, i.e., in addition to the links anticipated to emerge we also use data mining techniques to analyze the case that some of the existing links may probably be removed in the future. Both types of nodes which are anticipated to emerge or disappear are at least equally important to help domain expert in building rich understanding of the investigated domain.
In real life, social networks are highly dynamic objects; they grow and change quickly over time through the addition of new edges, signifying the appearance of new interactions in the underlying social structure (Nowell and Kleinberg 2003). Some challenging questions might arise, e.g., for how long a pair of entities will be connected together or can a link disappear? How about entities that are not connected in the current state, is it possible that they will get connected sometime in the future? How probable a link that disappeared recently will appear again in the future? How probable a link that recently emerged will disappear in the future? The ability to predict such changes with high accuracy is important to plan better for the future. For instance, the ability to predict how a business will grow or shrink may help in expanding facilities to absorb the growth or taking precautions to avoid the loss.
The problem of link prediction has attracted considerable attention in the research community, e.g., (Clauset et al. 2008; Dunlavy et al. 2011; Getoor and Diehl 2005; Tylenda et al. 2009). However, researchers concentrated mostly on predicting how a social network may grow by adding new links. In other words, most of the previous works on link prediction have either limited their research on the prediction of the links that will be added to the network during the interval from time t to a given future time \(t^{\prime}\) or implicitly have devoted the link prediction in specific domains such as co-authorship. Predicting links that may be dropped and shrinking a social network is still to be investigated.
This paper presents a novel model that tackles both emerging and shrinking problems. Formally, we define the link shrinking problem as following. Given a social network structure at time t, we seek to predict the status of the social network at time \(t^{\prime}\) by predicting the links that are expected to be added to or removed/dropped from the network. The model proposed in this paper is capable of equally predicting links that may appear as well as links that may disappear in the future. The model has been tested using two important applications from different yet interrelated domains, namely health care and gene expression networks. The reported results demonstrate the applicability and effectiveness of the proposed model.
The rest of the paper is organized as follows. Section 2 discusses background and our contributions on this work. Section 3 summarizes the related work in the area of link prediction. Section 4 presents and tests our model. The reported results are analyzed in Sect. 5. Finally, Sect. 6 concludes the paper with future directions.
2 Background and contributions
We have chosen two domains that are considered to be the most important domains that would concern people in the daily life, namely gene expression networks and health care representing two different structures one-mode and two-mode (known as bipartite) networks, respectively. Gene expression networks are extensively studied in the literature as part of the research effort to identify disease biomarkers. Biomarkers are particular molecules who behave in the cell different from their normal function. Drugs should be designed in a way to tackle such biomarkers in order to produce more effective results with minimum or no side effect. On the other hand, physicians in the health care domain do consult with each other in order to maximize the benefit from individual expertise and to combine personal experiences into a more robust and effective model of treatment to the benefit of patients. Studying interactions in both domains is crucial and could lead to novel and interesting discoveries.
A network is said to be two mode network or bipartite graph when it contains two different sets of nodes and no two nodes in the same set are linked together (Borgatti 1997). In our case, the referral network is considered to be a bipartite graph because we have sets of general practitioners (GPs) and specialists (SPs) and the links occur between a GP and an SP only. The one-mode network is a network that contains nodes of the same set. An example of the one-mode network will be a social network of students. Our link prediction approach differs from the other approaches described in literature that it can be applied to both network modes in addition to the other advantages that we will discuss in detail in the next section. Accordingly, we describe our contributions as to solve the two problems covered in the following subsections.
2.1 Gene expression network link prediction problem
Network analysis and prediction is an important problem domain tackled by different disciplines such as computer science and computational biology (Basso et al. 2005). The complex nature of gene expression data is often ascribed to its sensitivity and volatility: links are sensitive in that a small perturbation in expression values might indicate a crucial pattern or start of certain trend; and it is volatile due to noise and uncertainty. A key bottleneck in mining gene expression networks in current research is the difficulty to interpret gene–gene interactions with different timings.
In this paper, we propose a model based on link prediction to alleviate the aforementioned bottleneck and achieve the goal of predicting network interactions between genes. In this paper, we used the time series microarray data (DeRisi et al. 1997) from yeast and divide the entire experiment into two parts based on time intervals and inferred regulatory networks based on individual parts. Overall, our proposed method provides a social network-based model facilitating comparative gene–gene interaction analysis and effective visualization of network patterns. Predictions are illustratively made by linking and unlinking genes in networks. We do argue that genes and other molecules within the cell are analogous to humans (Naji et al. 2011) in the sense that they do form communities, interact and change camps after the internal structure of a molecule is mutated to make it more fit to a new activity which may disturb the whole biological system.
Problem formulation
Given time ordered some gene expression datasets for time intervals t A and t B , we intend to construct gene networks G t A = (V t A , E t A ) and G t B = (V t B , E t B ), where V t A and V t B represent set of genes with different time stamps and E t A and E t B represent connections between them based on some measure. We wish to predict links in G t B based on G t A .
2.2 The medical referral link prediction problem
Mining technique-based data analysis has recently received a significant attention in the healthcare industry; Healthcare data structure is too complex as it grows and researchers found that data mining techniques can analyze complex healthcare data and find patterns to improve the decision making process. As quoted in Koh and Tan (2005), “In healthcare, data mining is becoming increasingly popular, if not increasingly essential.” There are a lot of different data mining applications that can be applied to healthcare data. “The most common and important applications in data mining probably involve predicting models” (Koh and Tan 2005). In our paper, we focus on developing link prediction techniques that are capable of solving one of the most important problems in a highly crucial healthcare system called medical referral system as we will explain in details next.
The medical referral process is the process of referring patients to physicians with specific area of specialty. The model has been inspired from the health care system in Canada which works as illustrated in Fig. 1. When a patient has a problem, he/she visits a GP; if the GP cannot identify the problem of the patient, then the GP will refer the patient to a specialized physician referred as SP, who is known as specializing in the patient’s illness type.
As the healthcare delivery networks grows, the problem of finding the right specialist and communicating patient information from one provider to another becomes more difficult (Clancy et al. 1996; Lee et al. 1983). The reason behind the unacceptable delay in the referral process comes from the long time spent in finding the appropriate specialist. In addition, patients are sometimes referred to specialists who are busy and, hence, cannot accept further referrals for a specific period. GPs do not get quick reply for their inquiries regarding the SPs’ availability. This makes it even worse to wait for several weeks and then getting a response as not being able to accept additional referrals. In this paper, we form the social network of physicians associated with the medical referral process and then apply our link prediction model to predict which specialists are more likely to receive future referrals, and which specialists are more likely to lose their links with the GPs in the same network.
Problem formulation
Given two categories of physicians (GPs and SPs), we intend to construct a social network as a bipartite graph G = (V 1, V 2, E), where V 1 is GPs, V 2 is SPs and E is the set of connection between them reflecting the occurrence of a referral, i.e., if there is a link between a GP and an SP, it means that the SP has accepted a referral from the GP. Then, we seek to predict referral occurrences in the same social network constructed in a different time in the future.
3 Related work
The social network model has received considerable attention in the research community. It has been successfully adapted to investigate different applications such as terrorist networks (Ressler 2006), online communities (Crandall et al. 2008), and calling communities (Kianmehr and Alhajj 2009), among others. We argue that the model is powerful enough and can be used in analyzing any domain that could be represented in a network structure. Some other works have focused on link prediction issues. For example, the work described in Liu and Lu (2010) introduced a link prediction model based on the similarity of the nodes. This is important in applications that consider the similarity of nodes such as gender, age, etc. They proposed two similarity indices based on random walk. Other works apply link prediction in friendship applications where a person can be recommended to be a friend of another person based on mutual friends. The work described in Gilbert and Karahalios (2009) applied link prediction techniques to predict tie strength in social media. Shortcomings of these works are described as follows:
-
limitation to undirected networks where multiple or self-link directions are not allowed,
-
links are predicted based on one time period and not compared with how the prediction will be changed in different time periods using the same nodes,
-
predictions are limited to the addition of the links and ignore the removal of other links,
-
predictions are not studied based on the effect of node removal.
To the best of our knowledge, most of the previous works have concentrated on predicting links that might exist in the future (positive link prediction); there is no research effort described in the literature for predicting links that may disappear in the future (negative link prediction), though it is very important in determining the loss of relationships; and this may lead to weaken the network structure. In addition, no one has yet tackled the link prediction problem within the scope of gene expression networks and the health care domain which are considered as two of the most important domains for humans. One previous work related to our research has been done by Hasan et al. (2006) to study link prediction as a supervised learning task in the authorship domain. They identified a set of features that are key to this domain. With the help of these features, they were able to propose a classification model to predict a set of authors who will work together in the future.
Allali et al. (2011) has introduced a new approach in link prediction called internal links and weighted projection to predict links in bipartite graphs. This approach performs well in some social networks to predict links that will appear in the future but does not predict which link will be lost. We give more details about how this approach works later as we compare this approach to our new link prediction model. Several existing methods in mining gene expression data are described in the literature. Lee et al. (2006) introduced the fuzzy element in dealing with financial features. Gao et al. (2009) extended the fuzzy prediction method using the combination of rough and fuzzy sets in deriving association rules. Traditional approaches such as using support vector machines and others are discussed in Cao and Tay (2003).
Our proposed model will be tested on the gene expression networks and health care domains where we identify a set of features that we believe are powerful to build an accurate classification model for the link prediction purpose.
4 Model discussion and testing
Our model consists of two sections; predicting links to be added to the model, termed as positive link prediction (PLP) and predicting links to be dropped from the model, termed as negative link prediction (NLP). Figure 2 shows the steps taken to perform PLP and NLP. Next we explain each one of these steps in detail.
4.1 Positive link prediction
In the positive link prediction task, we seek to accurately predict future links given that they do not exist in the current network structure. We tested this model using data from the medical referral process and gene expression networks. This task is achieved by the following steps.
4.1.1 Data
Data should be collected for all the entities involved in the social network. Data can be collected in many different ways such as interviews, surveys, etc. We adopted the medical referral data from Anderson and Jay (1985) which is a matrix of 24 physicians (labeled A − X). For the gene expression networks, we have used different network centrality measures.
4.1.2 Preprocessing
Data is prepared by constructing an appropriate format that fits the experimental analysis. In our link prediction problem, we prepare the data in matrix format where rows represent GPs and columns represent SPs (for the medical referral problem) and matrix entries represent whether a GP made a referral to the corresponding SP or not (0 or 1, respectively).
4.1.3 Feature extraction
In the link prediction problem, a set of features have to be defined in order to be used in building the classification model. It is very important to choose the right features for any classifier. For the medical referral process, we have used the following features which have been derived from domain-specific information known mostly by common sense.
Ethnicity (E) This feature directly measures the proximity of a pair of nodes (a GP and an SP). Here we check each pair of doctors whether they come from the same ethnicity or not. Based on a survey done by the authors Anderson and Jay (1985), it has been concluded that if two doctors come from the same ethnicity, it is more likely that they know each other and hence they will have referrals associated between them.
Professional activity match (PAM) According to Anderson and Jay (1985), a group of physicians who are engaged in professional activities are more likely to have collaboration together. This feature will be very helpful in our classifier because if a new physician is engaged in a professional activity, he/she is more likely to have associations with other physicians involved in the same professional activity.
Sum of patients (SofP) The value of this feature is calculated by adding the number of patients that the pair of physicians have in the training years. The choice of this feature comes from the fact that physicians having higher patients count are more prolific. If either or both physicians are prolific, the probability is that this pair will collaborate is higher compared to the probability for the case of any random pair of physicians (Anderson and Jay 1985).
Sum of neighbors (SofN) This feature identifies the social connectivity of the pair of doctors, by adding the number of neighbors they have. In this feature, the neighborhood is obtained from the referral patterns information. The weight of this feature is the total number of the pair’s neighbors. Choosing this feature comes from the fact that if a GP refers a lot of patients and an SP receives a lot of patients, it is more likely that both doctors will have a relationship at some point in the future (Anderson and Jay 1985).
Jaccard similarity (JS) This feature requires an extended social network by adding the GPs consultations network. This means, a GP will have an additional link with another GP if one of them consults with the other. This feature is similar to the sum of neighbors feature by emphasizing that a pair of doctors may have common neighbors because each one has a lot of neighbors, not because they are strongly related to each other. The following Jaccard formula was used to compute the similarity:
The choice of this feature comes from the fact that if GP1 sends referrals to SP1 and then GP2 consults with GP1, it is more likely that GP1 will recommend SP1 to GP2. Based on the Jaccard formula, the more common GPs between x and y, the more likely they will be close to each other given the score is between zero and one, with one indicating the closest distance.
4.1.4 Classification
After the features are extracted, they are represented in a matrix form where rows mark data points whereas columns stand for feature numbers. The last column of the classification data represents the class label. Then the matrix is used as input to selected classifiers in order to perform training and testing. For the PLP of the medical referral data, we have listed the five features discussed above as the columns of the classification dataset and each row represent a pair of GP and SP. The class label is 0 or 1 representing whether a link (referral) exists between the pair of physicians in the original referral matrix or not. On the other hand, for the gene expression dataset, we have used the centrality measures described above as the columns.
4.2 Negative link prediction
The goal of this model is the opposite of PLP’s goal, where we seek in this stage to accurately predict links that are more likely to be removed from the network sometime in the future.
4.2.1 Do PLP
To perform the NLP for any data, we have to apply the PLP steps first so that we can use the social network from the output of the PLP as the input to the NLP, in addition to the comparison purposes.
4.2.2 NLP algorithm
Once we have the final matrix (referred by the original matrix) from the PLP, we apply the following algorithm:
-
Step 1 Invert the original matrix; by inverting we mean replace 0 by 1 and vice versa.
-
Step 2 Predict the new links of the inverted matrix using PLP.
-
Step 3 Invert the new matrix resulted from Step 2 (modified matrix)
-
Step 4 Discover which links are reported to be missing compared to the original matrix.
We have tested the NLP method using the medical referral data. Next, we report and discuss testing results of both domains.
5 Results and discussions
5.1 Success criteria
The success criteria are expressed in terms of percentages of accuracy for the PLP and in terms of graph statistics for the NLP. Accuracy is the ratio of the sum of all data points classified correctly over the number of all data points.
5.2 Medical referral testing
Figure 3 shows the classification accuracy used for the PLP model on the medical referral dataset. It shows that the SVM algorithm has reported the highest accuracy (92 %). The great success of the future links reported by our classifier show how important the features we have selected in the classification dataset are. Figure 4 shows the network of the original referral matrix between GPs (round nodes) and SPs (square nodes). A link between a GP and an SP means that the GP sent referral to the SP. Figure 4b shows the resulting network as reported by our classifier. Links in green are the links anticipated to be added in the future. Figure 4c shows the resulting network of the potential links (in red) to be removed from the original graph in the future as reported by the classifier. Figure 4d shows links reported to be added to the original network after removing the central node.
Table 1 summarizes the statistics of the three results reported in Fig. 4a–d. The first column of the table lists the SP nodes. Original number of links field lists the number of links associated with each corresponding SP. Links added field shows the number of predicted links to be added by our proposed PLP model. Links removed field displays the number predicted links to be removed by our NLP model. Links added then removed and links removed then added fields show the numbers of links which were predicted to be added by our PLP then were removed by our proposed NLP and vice versa. The last field shows the links predicted to be added after removing the most significant specialist node from the network. From this table we conclude the following:
Links added the potential links predicted to be added do not depend on how strong the node is in the network, but rather depend on how the links are involved in the network. For example, J, T, and U are almost ignored in the network, but have gained links which have increased and enforced their participation in the network. This also tells us that having some current information about a pair of doctors will allow us to find which specialist will be the busiest in the future by simply counting the new total number of links for each SP. This will be very helpful to the health referral system where it will allow the GPs to avoid referring patients to those SPs who are reported as would be potentially busy; and this will help in avoiding the referrals delay problem in advance. Moreover, once we know the SPs that are predicted to be busy in the future, we can seek recommendations from them for alternative specialist(s) to refer patients to. Finally, such information may be valuable to plan better to be ready to deal with the predicted epidemics which may be anticipated if certain type of links is heavily introduced in the model like more people visiting pediatricians may be an indicator of epidemic affecting the children.
Links removed this is very important to understand the loss of relations and how it affects the structure of the network. If a GP has one link and this link is going to be dropped in the future, it will make this GP isolated from the network which should not happen. This feature is also used to support and validate the predictions of the potential links to be added; whether these potential links are predicted to be removed or not. From Table 1, we see that nodes O, T, J, and K have small number of links, and none of their links was detected as potential to be removed; this is true and valid as supported by having many green links predicted for them. Moreover, the location of L is weaker than most of the others as it has only 5 original links. Our model predicted to add 3 more links to this node to make it more engaging in the network to support the interaction of the community.
Links added then removed and links removed then added We use this feature to test the prediction decisions made based on the previous features. In other words, we check if any of the links that has been predicted to be added are predicted to be removed and vice versa. As reported in Table 1, none of the links have been added is predicted to be removed and vice versa; this means the predictions potentially reflect good anticipation of the future.
Links added after removing the most important node in the health care domain, it happens that an SP leaves the practice by retiring or may move. The network structure definitely changes to reflect the latter situation. To study this experimentally, we have deleted the most central SP node from the original graph and applied the link prediction model to see how the links will be distributed in the network. We have noticed that those SPs who have few links have gained new links. Our interpretation of this could be articulated as follows: those SPs with few links did not receive referrals because they were not as popular as the node we removed. But, when the most popular node was deleted those new SPs received new links to substitute the links that were connected to the deleted node.
5.3 Gene expression testing
The PLP has also been tested on a real-world gene expression dataset (DeRisi et al. 1997). We constructed the regulatory network from the gene expression data using the ARACNE algorithm (Basso et al. 2005), which is a robust reverse engineering algorithm using mutual information between co-expression values of genes. The dataset consists of 100 gene expressions at four different time snapshots. First, we have constructed the gene expression network where a link between two genes means both genes have an expression value above the average expression value of the whole dataset. Second, we constructed a classification dataset using seven network features: Centrality Authority, Centrality Closeness, Centrality Column Degree, Centrality Eigenvector, Centrality in Closeness, Centrality in Degree, and Centrality in Degree. Rows of the classification dataset are pairs of all possible genes in the network. The values of each one of these features are between 0 and 1. Third, we have constructed a class label with the value of “P” or “N” to reflect if the link currently exists or not, respectively. Finally, we have used three different classifiers to classify whether a link between two genes will appear in the next time snapshot or no given that this link does not currently exist. Figure 5 shows the accuracy of the classifiers.
5.4 Internal links approach versus PLP
The link prediction model using internal links and weighted projection introduced in Allali et al. (2011) plays an important role in bipartite graphs. However, it does not perform well for health care social networks. The internal link method depends on searching for links that are classified as internal links. If any internal link is found, it is considered as a potential link to appear in the future. As defined in Allali et al. (2011), a link is said to be considered as internal if and only if the links it induces already exist in the projection of the network. We have applied this method with no threshold to predict the most possible links to our referral network and below we give the results.
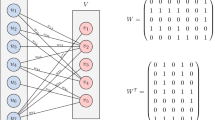
Figure 6 shows the projection of the referral network. It contains nodes of GPs linked together if they have a common neighbor. Neighbors are in this case SPs. Based on the internal link prediction, in order for a link to be predicted in the original network, all the links that this link induces must exist in the projection graph. For example, in Fig. 7, there is no link currently between nodes b and A. Adding a link between them will induce pairs of {b, c}, {b, d}, {b, g}, {b, i}, {b, j}, {b, n}, and {b, u}. All these pairs are linked in the projection graphs. Therefore, we consider the link between b and A as an internal link that is predicted to appear in the future.
We counted the number of links expected to be added to every SP in the referral network using our approach and the internal links approach. The results are shown in Figs. 8 and 9. We argue that our approach has more advantages because of the following reasons:
As shown in Fig. 8, internal link prediction resulted in spreading the communities of the SPs and created too long distances between the data points which cause the scattered or not well-distributed clusters. Instead, our results show very well-clustered data points. The goal of data mining in social networks, especially referral data, is to build well-organized communities and make all nodes evenly active to control the referral traffic between GPs and SPs.
The number of predicted links is not distributed in a way to control the traffic of the referrals. For example, specialists J, N, and U receive the highest number of referrals which will increase the referral wait time. In addition, node K received 8 links using the internal links prediction and received nothing using PLP. In the referral system, we believe that node K is connected with 11 GPs already which send referrals frequently to K. Therefore, we should reduce the amount of referral traffic to this SP in order not to add significant GP ties to it.
6 Conclusions and future directions
In the link prediction field, this work is considered to be the first model to consider predicting negative links. We have shown how our model can predict the links that have potential to be removed from the network. In the domain of health care and gene expression networks, this work can be considered to be the first step towards building a prediction model to predict future associations between physicians; it can be very helpful to reduce the referral traffic. In addition, our proposed link prediction model has successfully predicted the co-expression values of genes in the network setting. We compared our PLP approach with the internal links approach and we concluded that our approach produced results with better interpretation in favor to the referral data. We are currently applying the same model to other domains including terrorist networks, financial data and employees’ networks in order to illustrate the significance of the proposed model and its applicability to various domains. Future directions will focus on including patient’s data in order to see how it will affect the link prediction decision. We believe that including patient’s data will result in more accurate link predictions leading to a better decision making process. In addition, our future plan includes using more classification algorithms to enhance the PLP model.
References
Allali O, Magnien C, Latapy M (2011) Link prediction in bipartite graphs using internal links and weighted projection. In: 2011 IEEE conference on computer communications workshops (INFOCOM WKSHPS), pp 936–941
Anderson JG, Jay SJ (1985) Computers and clinical judgement: the role of physician networks. Soc Sci Med 20(10):969–979
Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A (2005) Reverse engineering of regulatory networks in human B cells. Nat Genet 37(4):382–390
Borgatti S (1997) Network analysis of 2-mode data. Soc Netw 19(3):243–269
Cao L, Tay F (2003) Support vector machine with adaptive parameters in financial time series forecasting. IEEE Trans Neural Netw 14(6):1506–1518
Clancy CM, Lanier D, Grady ML (1996) Research at the interface of primary and specialty care. AHCPR 96-0034
Clauset A, Moore C, Newman MEJ (2008) Hierarchical structure and the prediction of missing links in networks. Nature 453(7191):98–101
Crandall D, Cosley D, Huttenlocher D, Kleinberg J, Suri S (2008) Feedback effects between similarity and social influence in online communities. In: Proceeding of the 14th ACM SIGKDD international conference on Knowledge discovery and data mining. ACM, New York, pp 160–168
DeRisi JL, Iyer VR, Brown PO (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686
Dunlavy DM, Kolda TG, Acar E (2011) Temporal link prediction using matrix and tensor factorizations. ACM Trans Knowl Discov Data (TKDD) 5(2), Art no 10
Gao S, Alhajj R, Rokne JG (2009) Modeling knowledge discovery in financial forecasting. In: Proceedings of the IEEE International Conference on Information Reuse and Integration, pp 41–46
Getoor L, Diehl CP (2005) Link mining: a survey. SIGKDD Explor Newsl 7(2):3–12
Gilbert E, Karahalios K (2009) Predicting tie strength with social media. In: Proceedings of the 27th international conference on Human factors in computing systems, CHI ’09. ACM, New York, pp 211–220
Hasan MA, Chaoji V, Salem S, Zaki M (2006) Link prediction using supervised learning. In: Proceedings of SDM 06 workshop on link analysis, counterterrorism and security, pp 936–941
Kianmehr K, Alhajj R (2009) Calling communities analysis and identification using machine learning techniques. Expert Syst Appl 36(3, Part 2):6218–6226
Koh HC, Tan G (2005) Data mining applications in healthcare. J Healthc Inf Manag 19(2):64–72
Lee CHL, Liu A, Chen WS (2006) Pattern discovery of fuzzy time series for financial prediction. IEEE Trans Knowl Data Eng 18(5):613–625
Lee T, Pappius EM, Goldman L (1983) Impact of inter-physician communication on the effectiveness of medical consultations. Am J Med 74(1):106–112
Liu W, Lu L (2010) Link prediction based on local random walk
Naji G, Nagi M, Elsheikh AM, Gao S, Kianmehr K, Özyer T, Demetrick D, Alhajj R, Rokne J, Ridley M (2011) Effectiveness of social networks for studying biological agents and identifying cancer biomarkers. In: Counterterrorism and open source intelligence, pp 285–313
Nowell DL, Kleinberg J (2003) The link prediction problem for social networks. In: Proceedings of the twelfth international conference on Information and knowledge management, CIKM ’03. ACM, New York, pp 556–559
Ressler S (2006) Social network analysis as an approach to combat terrorism: past, present, and future research. Homel Secur Aff 2(2). http://www.hsaj.org
Tylenda T, Angelova R, Bedathur S (2009) Towards time-aware link prediction in evolving social networks. In: Proceedings of the 3rd workshop on social network mining and analysis, SNA-KDD ’09. ACM, New York, pp 1–10
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Almansoori, W., Gao, S., Jarada, T.N. et al. Link prediction and classification in social networks and its application in healthcare and systems biology. Netw Model Anal Health Inform Bioinforma 1, 27–36 (2012). https://doi.org/10.1007/s13721-012-0005-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13721-012-0005-7