Abstract
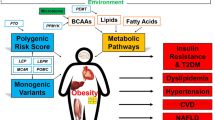
Cardiometabolic dieases are the main causes of morbidity and mortality worldwide. Despite large clinical and basic research efforts to improve the understanding of the molecular mechanisms of these conditions and to develop prediagnostic markers still remains a challenge. Recently, targeted and untargeted metabolomics technique became available for comprehensive studies of small molecules in biological samples in large-scale studies. MS- or NMR-based techniques dominate and have recently been used extensively in studies to identify predictive biomarkers, monitoring therapeutic response as well as in basic mechanism studies of obesity, metabolic syndrome, type 2 diabetes, and heart disease. Here, we review the recent findings from such studies. Findings highlight the role of branched-chain amino acids, acyl carnitines and other lipid classes in these conditions. Putative biomarkers for disease conditions have been identified but mechanistic understanding of their role in the disease development and progression typically remains to be elucidated. Future studies should to a greater extent be designed to allow studies on causal mechanistic links between metabolites and disease.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Eckel RH, Kahn R, Robertson RM, Rizza RA. Preventing cardiovascular disease and diabetes: a call to action from the American Diabetes Association and the American Heart Association. Circulation. 2006;113(25):2943–6.
C Unachukwu SO. Diabetes mellitus and cardiovascular risk. The Internet Journal of Endocrinology. 2012. p. 1–10.
Upadhyay RK. Emerging risk biomarkers in cardiovascular. 2015;2015.
Castro JP, El-Atat FA, McFarlane SI, Aneja A, Sowers JR. Cardiometabolic syndrome: pathophysiology and treatment. Curr Hypertens Rep United States. 2003;5(5):393–401.
O’Flaherty M, Buchan I, Capewell S. Contributions of treatment and lifestyle to declining CVD mortality: why have CVD mortality rates declined so much since the 1960s? Heart England. 2013;99(3):159–62.
Basak T, Varshney S, Akhtar S, and Sengupta S. Understanding different facets of cardiovascular diseases based on model systems to human studies: a proteomic and metabolomic perspective. J Proteomics. 2015.
Milburn M V, Lawton KA, Mcdunn JE, Ryals JA, and Guo L. Genetics meets metabolomics. Suhre K, editor. New York, NY: Springer New York; 2012;177–90.
Scalbert A, Brennan L, Manach C, Andres-lacueva C, Dragsted LO, Draper J, Rappaport SM, Hooft JJJ Van Der, and Wishart DS. Narrative review. The food metabolome : a window over dietary exposure. 1 – 3. 2014;(C).
Moco S, Ross A. Can we use metabolomics to understand changes to gut microbiota populations and function? A nutritional perspective. In: Kochhar S, Martin F-P, editors. Metabonomics Gut Microbiota Nutr Dis SE - 5. London: Springer; 2015. p. 83–108.
Shulaev V. Metabolomics technology and bioinformatics. Briefings in Bioinformatics. 2006. p. 128–39.
Hedrick VE, Dietrich AM, Estabrooks PA, Savla J, Serrano E, Davy BM. Dietary biomarkers: advances, limitations and future directions. Nutr J Nutr J. 2012;11(1):109.
Ryan EP, Heuberger AL, Broeckling CD, Borresen EC, Tillotson C, Prenni JE. Advances in Nutritional Metabolomics. Curr Metabol. 2013;1:109–20.
Zhang A, Sun H, Wang P, Han Y, and Wang X. Modern analytical techniques in metabolomics analysis. The Analyst. 2012. p. 293.
Andersen M-BS, Rinnan A, Manach C, Poulsen SK, Pujos-Guillot E, Larsen TM, Astrup A, and Dragsted LO. Untargeted metabolomics as a screening tool for estimating compliance to a dietary pattern. J Proteome Res. 2014.
Emwas A-HM, Salek RM, Griffin JL, Merzaban J. NMR-based metabolomics in human disease diagnosis: applications, limitations, and recommendations. Metabolomics. 2013;9(5):1048–72.
Bajad S, Shulaev V. LC-MS-based metabolomics. Methods Mol Biol. 2011;708:213–28.
Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, et al. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Anal Chem. 2006;78(2):567–74.
Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–83.
Rappaport SM, Barupal DK, Wishart D, Vineis P, and Scalbert A. The blood exposome and its role in discovering causes of disease. Environmental Health Perspectives. Public Health Services, US Dept of Health and Human Services; 2014. p. 769–74.
Wientzek A, Tormo Díaz MJ, Castaño JMH, Amiano P, Arriola L, Overvad K, Østergaard JN, Charles MA, Fagherazzi G, Palli D, et al. Cross-sectional associations of objectively measured physical activity, cardiorespiratory fitness and anthropometry in european adults. Obesity. Nature Publishing Group; 2014;22(5).
Szymańska E, Bouwman J, Strassburg K, Vervoort J, Kangas AJ, Soininen P, et al. Gender-dependent associations of metabolite profiles and body fat distribution in a healthy population with central obesity: towards metabolomics diagnostics. OMICS. 2012;16(12):652–67.
World Health Organization. Global Status Report On Noncommunicable Diseases 2014. 2014.
Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A Branched-Chain Amino Acid-Related Metabolic Signature that Differentiates Obese and Lean Humans and Contributes to Insulin Resistance. Cell Metab. 2009;9(4):311–26.
Moore SC, Matthews CE, Sampson JN, Stolzenberg-Solomon RZ, Zheng W, Cai Q, et al. Human metabolic correlates of body mass index. Metabolomics. 2014;10(2):259–69.
Xie G, Ma X, Zhao A, Wang C, Zhang Y, Nieman D, et al. The Metabolite Profiles of the Obese Population Are Gender-Dependent. J Proteome Res. 2014;13(9):4062–73. Whereas circulating branched chain amino acid (BCAA) concentrations are frequently reported as elevated in obese compared to lean, three different case-control settings showed elevated BCAA only in obese men but not in obese women. The significance of this finding is unclear, but BCAA dysregulation may be more related to metabolic health status that obesity per se.
Mihalik SJ, Michaliszyn SF, De Las HJ, Bacha F, Lee S, Chace DH, et al. Metabolomic profiling of fatty acid and amino acid metabolism in youth with obesity and type 2 diabetes: Evidence for enhanced mitochondrial oxidation. Diabetes Care. 2012;35(3):605–11.
Wahl S, Yu Z, Kleber M, Singmann P, Holzapfel C, He Y, et al. Childhood obesity is associated with changes in the serum metabolite profile. Obes Facts. 2012;5(5):660–70.
Mccormack SE, Shaham O, Mccarthy MA, Deik AA, Wang TJ, Gerszten RE, et al. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr Obes. 2013;8(1):52–61.
Oberbach A, Blüher M, Wirth H, Till H, Kovacs P, Kullnick Y, et al. Combined proteomic and metabolomic profiling of serum reveals association of the complement system with obesity and identifies novel markers of body fat mass changes. J Proteome Res. 2011;10(10):4769–88.
Kim MJ, Yang HJ, Kim JH, Ahn CW, Lee JH, Kim KS, and Kwon DY. Obesity-related metabolomic analysis of human subjects in black soybean peptide intervention study by ultraperformance liquid chromatography and quadrupole-time-of-flight mass spectrometry. J Obes. 2013;2013.
Martin F-PJ, Montoliu I, Collino S, Scherer M, Guy P, Tavazzi I, et al. Topographical body fat distribution links to amino acid and lipid metabolism in healthy non-obese women. PLoS ONE. 2013;8, e73445. Differentiation between subtypes of adiposity (visceral and surface) in women revealed different metabolic profiles, where visceral adiposity is the subtype more associated with metabolic dysregulation.
Böhm A, Halama A, Meile T, Zdichavsky M, Lehmann R, Weigert C, Fritsche A, Stefan N, Königsrainer A, Häring HU, et al. Metabolic signatures of cultured human adipocytes from metabolically healthy versus unhealthy obese individuals. PLoS One. Public Library of Science; 2014;9(4). In vitro investigation of cultured adipocytes from insulin resistant vs insulin sensitive obese subjects suggested that intracellular/extracellular dysregulation of phospholipids may be more related to metabolic health status that obesity per se.
Kim JY, Park JY, Kim OY, Ham BM, Kim HJ, Kwon DY, et al. Metabolic profiling of plasma in overweight/obese and lean men using ultra performance liquid chromatography and Q-TOF Mass spectrometry (UPLC-Q-TOF MS). J Proteome Res. 2010;9(9):4368–75.
Wang C, Feng R, Sun D, Li Y, Bi X, Sun C. Metabolic profiling of urine in young obese men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC/Q-TOF MS). J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879(27):2871–6. Elsevier BV.
Calvani R, Miccheli A, Capuani G, Tomassini Miccheli A, Puccetti C, Delfini M, et al. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int J Obes (Lond). 2010;34(6):1095–8. Nature Publishing Group.
International Diabetes Federation. The IDF consensus worldwide definition of the metabolic syndrome. The IDF consensus worldwide definition of the metabolic syndrome. 2006.
Kaur J. A comprehensive review on metabolic syndrome. Cardiol Res Pract. 2014;ID 943162.
Oda E. Metabolic syndrome: Its history, mechanisms, and limitations. Acta Diabetol. 2012;49(2):89–95.
Eckel RH, Alberti KGMM, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2010;375(9710):181–3.
Grundy SM. Metabolic syndrome: a multiplex cardiovascular risk factor. J Clin Endocrinol Metab. 2007;92(2):399–404.
Galassi A, Reynolds K, He J. Metabolic syndrome and risk of cardiovascular disease: a meta-analysis. Am J Med. 2006;119(10):812–9.
Grundy SM, Brewer HB, Cleeman JI, Smith SC, Lenfant C. Definition of Metabolic Syndrome: Report of the National Heart, Lung, and Blood Institute/American Heart Association Conference on Scientific Issues Related to Definition. Circulation. 2004;109(3):433–8.
Chen S, Chu Y, Zhao X, Gao P, Zhang L, Zhan L, et al. HPLC-MS-Based Metabonomics Reveals Disordered Lipid Metabolism in Patients with Metabolic Syndrome. J Anal Sci Technol. 2011;2(Supplement A):A173–8.
Huang CF, Cheng ML, Fan CM, Hong CY, Shiao MS. Nicotinuric acid: A potential marker of metabolic syndrome through a metabolomics-based approach. Diabetes Care. 2013;36(6):1729–31.
Lin Z, Vicente Gonçalves CM, Dai L, Lu HM, Huang JH, Ji H, et al. Exploring metabolic syndrome serum profiling based on gas chromatography mass spectrometry and random forest models. Anal Chim Acta. 2014;827:22–7. Elsevier BV.
J.H Hong H, Eun Y, Hee J, Soon K, and Jin H. Urinary chiro- and myo-inositol levels as a biological marker for type 2 diabetes mellitus. 2012;33:193–9.
Gall WE, Beebe K, Lawton K a., Adam KP, Mitchell MW, Nakhle PJ, Ryals J a., Milburn M V., Nannipieri M, Camastra S, et al. Α-Hydroxybutyrate Is an Early Biomarker of Insulin Resistance and Glucose Intolerance in a Nondiabetic Population. PLoS One. 2010;5(5).
Yu Z-R, Ning Y, Yu H, Tang N-J. A HPLC-Q-TOF-MS-based urinary metabolomic approach to identification of potential biomarkers of metabolic syndrome. J Huazhong Univ Sci Technol Med Sci. 2014;34(2):276–83.
Dunn WB. Diabetes - the Role of Metabolomics in the Discovery of New Mechanisms and Novel Biomarkers. Curr Cardiovasc Risk Rep. 2012;7(1):25–32.
Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2010;365(9467):1333–46.
DeFronzo RA. Pathogenesis of type 2 diabetes mellitus. Med Clin N Am. 2004;88(4):787–835. ix.
Zheng Y, Hu FB. Comprehensive Metabolomic Profiling of Type 2 Diabetes. Clin Chem. 2015;61(3):453–5.
Lowe WL, Bain JR. “Prediction is very hard, especially about the future”: new biomarkers for type 2 diabetes? Diabetes. 2013;62(5):1384–5.
Suhre K, Meisinger C, Döring A, Altmaier E, Belcredi P, Gieger C, et al. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS ONE. 2010;5(11), e13953.
Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–53.
Wang-sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, Heim K, Meisinger C, Gieger C, Prehn C, et al. Novel biomarkers for pre-diabetes identified by metabolomics. 2012;(615).
Menni C, Fauman E, Erte I, Perry JRB, Kastenmüller G, Shin SY, et al. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes. 2013;62(12):4270–6.
Wang TJ, Ngo D, Psychogios N, Dejam A, Larson MG, Vasan RS, et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest. 2013;123(10):4309–17. In this study, 2-Aminoadipic acid was discorvered as a novel biomarker that predicted development of diabetes in normoglycemic individuals.
Floegel A, Stefan N, Yu Z, Mühlenbruch K, Drogan D, Joost HG, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62(2):639–48. This study included three independent, well-described study populations and links lifestyle and dietary patterns to a metabolic pattern associated with type 2 diabetes.
Padberg I, Peter E, González-Maldonado S, Witt H, Mueller M, Weis T, Bethan B, Liebenberg V, Wiemer J, Katus H a., et al. A new metabolomic signature in type-2 diabetes mellitus and its pathophysiology. PLoS One. 2014;9(1). This study combined data from different platforms and was the first that investigated if glyoxylate could be useful for prediction of of diabetic complications.
Liu L, Wang M, Yang X, Bi M, Na L, Niu Y, et al. Fasting serum lipid and dehydroepiandrosterone sulfate as important metabolites for detecting isolated postchallenge diabetes: Serum metabolomics via ultra-high-performance LC-MS. Clin Chem. 2013;59(9):1338–48.
Li X, Xu Z, Lu X, Yang X, Yin P, Kong H, et al. Comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry for metabonomics: Biomarker discovery for diabetes mellitus. Anal Chim Acta. 2009;633(2):257–62.
Zhang AH, Sun H, Yan GL, Yuan Y, Han Y, Wang XJ. Metabolomics study of type 2 diabetes using ultra-performance LC-ESI/quadrupole-TOF high-definition MS coupled with pattern recognition methods. J Physiol Biochem. 2014;70(1):117–28.
Drogan D, Dunn WB, Lin W, Buijsse B, Schulze MB, Langenberg C, et al. Untargeted metabolic profiling identifies altered serum metabolites of type 2 diabetes mellitus in a prospective, nested case–control study. Clin Chem. 2014;497:487–97. This study is one of the largest prospective metabolite profiling studies to find biomarkers of type 2 diabetes to date. A positive correlation of hydroxymethyl butenyl disphosphate and isopentenyladenosine-5’- monophosphatewith incident T2D was shown for the first time, leading to new hypotheses about T2D pathophysiology.
Tuomilehto J, Lindström J, Eriksson JG, Valle TT, Hämäläinen H, Ilanne-Parikka P, Keinänen-Kiukaanniemi S, Laakso M, Louheranta A, Rastas M, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. The New England journal of medicine. 2001.
Landberg R, Marklund M, Kamal-Eldin A, Åman P. An update on alkylresorcinols—occurrence, bioavailability, bioactivity and utility as biomarkers. J Funct Foods. 2014;7(1):77–89. Elsevier Ltd.
Floegel A, von Ruesten A, Drogan D, Schulze MB, Prehn C, Adamski J, et al. Variation of serum metabolites related to habitual diet: a targeted metabolomic approach in EPIC-Potsdam. Eur J Clin Nutr. 2013;67(10):1100–8. Nature Publishing Group. Nature Publishing Group. This study linked dietary patterns characterized by intakes of red meat, whole-grain bread, tea and coffee with chronic disease relevant metabolites.
Floegel A, Wientzek A, Bachlechner U, et al. Linking diet, physical activity, cardiorespiratory fitness and obesity to serum metabolite networks: findings from a population-based study. Int J Obes. 2014;38:1388--96.The association between phenotypes and lifestyle characteristics with metabolites networks was invetigated by using an innovative network approach in a large cohort study.
Guertin KA, Moore SC, Sampson JN, Huang WY, Xiao Q, Stolzenberg-Solomon RZ, et al. Metabolomics in nutritional epidemiology: Identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am J Clin Nutr. 2014;100(1):208–17.
Cooper AJ, Sharp SJ, Luben RN, Khaw K-T, Wareham NJ, Forouhi NG. The association between a biomarker score for fruit and vegetable intake and incident type 2 diabetes: the EPIC-Norfolk study. Eur J Clin Nutr. 2015;69(4):449–54. Nature Publishing Group.
Wittenbecher C, Muhlenbruch K, Kroger J, Jacobs S, Kuxhaus O, Floegel A, et al. Amino acids, lipid metabolites, and ferritin as potential mediators linking red meat consumption to type 2 diabetes. Am J Clin Nutr. 2015;101(6):1241–50. This is the first study evaluating a large set of metabolites as potential dietary biomarkers of red meat intake and and associated it with a metabolomic risk profile for type 2 diabetes.
Mendis S, Puska P, and Norrving B. Global atlas on cardiovascular disease prevention and control. World Heal Organ. 2011;2–14.
Rasmiena AA, Ng TW, Meikle PJ. Metabolomics and ischaemic heart disease. Clin Sci (Lond). 2013;124(5):289–306.
Luan H, Chen X, Zhong S, Yuan X, Meng N, Zhang J, et al. Serum metabolomics reveals lipid metabolism variation between coronary artery disease and congestive heart failure: a pilot study. Biomarkers. 2013;18(4):314–21.
Kang SM, Park JC, Shin MJ, Lee H, Oh J, do Ryu H, et al. (1)H nuclear magnetic resonance based metabolic urinary profiling of patients with ischemic heart failure. Clin Biochem. 2011;44(4):293–9.
Shah SH, Granger CB, Hauser ER, Kraus WE, Sun JL, Pieper K, et al. Reclassification of cardiovascular risk using integrated clinical and molecular biosignatures: Design of and rationale for the Measurement to Understand the Reclassification of Disease of Cabarrus and Kannapolis (MURDOCK) Horizon 1 Cardiovascular Disease. Am Heart J Mosby, Inc. 2010;160(3):371–9.e2.
Shah SH, Sun J-L, Stevens RD, Bain JR, Muehlbauer MJ, Pieper KS, et al. Baseline metabolomic profiles predict cardiovascular events in patients at risk for coronary artery disease. Am Heart J Mosby, Inc. 2012;163(5):844–50.e1.
Shah SH, Bain JR, Muehlbauer MJ, Stevens RD, Crosslin DR, Haynes C, et al. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet. 2010;3(2):207–14.
Stegemann C, Pechlaner R, Willeit P, Langley SR, Mangino M, Mayr U, et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based bruneck study. Circulation. 2014;129(18):1821–31.
Rizza S, Copetti M, Rossi C, Cianfarani MA, Zucchelli M, Luzi A, et al. Metabolomics signature improves the prediction of cardiovascular events in elderly subjects. Atherosclerosis. 2014;232(2):260–4. Elsevier Ltd.
Ganna A, Salihovic S, Sundström J, Broeckling CD, Hedman ÅK, Magnusson PKE, et al. Large-scale metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS Genet. 2014;10(12), e1004801. The largest untargeted metabolomics study to find biomarkers for coronary heart disease and to delineate the underlying mechanisms and causal effects of novel biomarkers detected in more than 3600 individuals.
Jiang Z, Sun J, Liang Q, Cai Y, Li S, Huang Y, et al. A metabonomic approach applied to predict patients with cerebral infarction. Talanta. 2011;84(2):298–304. Elsevier BV.
Jung JY, Lee H-S, Kang D-G, Kim NS, Cha MH, Bang O-S, et al. 1H-NMR-based metabolomics study of cerebral infarction. Stroke. 2011;42(5):1282–8.
Kimberly WT, Wang Y, Pham L, Furie KL, Gerszten RE. Metabolite profiling identifies a branched chain amino acid signature in acute cardioembolic stroke. Stroke. 2013;44(5):1389–95.
Jove M, Mauri-Capdevila G, Suarez I, Cambray S, Sanahuja J, Quilez A, et al. Metabolomics predicts stroke recurrence after transient ischemic attack. Neurol. 2015;84(1):36–45. United States.
Beatty AL, Ku IA, Bibbins‐Domingo K, Christenson RH, DeFilippi CR, Ganz P, et al. Traditional Risk Factors Versus Biomarkers for Prediction of Secondary Events in Patients With Stable Coronary Heart Disease: From the Heart and Soul Study. J Am Heart Assoc. 2015;4(7), e001646.
Dumas ME, Kinross J, and Nicholson JK. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and fatty liver disease. Gastroenterology. 2014. p. 46–62.
Putri SP, Nakayama Y, Matsuda F, Uchikata T, Kobayashi S, Matsubara A, and Fukusaki E. Current metabolomics: Practical applications. Journal of Bioscience and Bioengineering. 2013. p. 579–89.
Johnson CH, Ivanisevic J, Benton HP, Siuzdak G. Bioinformatics: the next frontier of metabolomics. Anal Chem. 2014;87(1):147–56.
Kell DB. Systems biology, metabolic modelling and metabolomics in drug discovery and development. Drug Discovery Today. 2006. p. 1085–92.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Carl Brunius, Lin Shi, and Rikard Landberg declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Public Health and Translational Medicine
Carl Brunius and Lin Shi contributed equally to this work.
Rights and permissions
About this article
Cite this article
Brunius, C., Shi, L. & Landberg, R. Metabolomics for Improved Understanding and Prediction of Cardiometabolic Diseases—Recent Findings from Human Studies. Curr Nutr Rep 4, 348–364 (2015). https://doi.org/10.1007/s13668-015-0144-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13668-015-0144-4