Abstract
Two dimeric abietane diterpenoids, salviwardins A and B (1 and 2), and a seco-abietane diterpenoid salviwardin C (3), along with five known analogues (4–8), were isolated from the roots of Salvia wardii. The structures of these isolates were elucidated by extensive spectroscopic methods. The inhibitory activities of these isolates against five human cancer cell lines in vitro were also tested.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The genus Salvia is a rich source of diterpenoids with structural diversity [1–4]. Hundreds of diterpenoids with interesting bioactivities, such as tanshinone IIA (treat cardiovascular diseases), salvicine (a significant antitumor agent), neotanshinlactone (inhibition of breast cancer), and salvinorin A (the first non-nitrogenated opium receptor agonist), have been characterized from the plants within this genus [5–8]. Many species of this genus, such as S. miltiorrhiza, S. yunnanensis, and S. przewalskii, are used to treat cardiovascular diseases [9–11], and S. prionitis is used in Chinese folk medicine for the treatment of tonsillitis, pharyngitis, and bacillary dysentery [12].
Salvia wardii, a herb with violet flowers distributed in east of Tibet, has not been chemically studied before [13]. Aiming at searching for structurally interesting and bioactive diterpenoids from the Salvia plants, we chemically investigated S. wardii and isolated three new abietane diterpenoids, salviwardins A–C (1–3), and five known analogues (4–8). The inhibitory activities of these isolates against five human cancer cell lines in vitro were also tested.
2 Results and Discussion
The acetone extract of the air-dried and powdered the roots of S. wardii (33 kg) was subjected to a silica gel column to afford fractions A–G. Fraction B was subjected to a series of chromatographic methods, and led to the isolation of three new abietane derivatives, salviwardins A–C (1–3), together with five knows analogues, including prionitin (4) [14], sahandol (5) [15], salvilenone (6) [16], microstegiol (7) [17], and ferruginol (8) [18].
Salviwardin A (1) was obtained as orange powder. Its molecular formula C40H54O4 was established by its 13C NMR and HREIMS (m/z 598.4029, [M]+) data, indicating 14 degrees of unsaturation. The IR absorption at 3440 and 1624 cm−1 implied the existence of hydroxyl and carbonyl groups. The 13C and DEPT NMR (Table 1) spectroscopic data of 1 revealed 40 carbon signals, comprising fifteen quaternary carbons (one carbonyl, nine olefinic, and one oxygenated group), seven methines (one oxygenated and three olefinic ones), eight methylenes and ten methyls. The 1H NMR (Table 2) spectrum of 1 showed the presence of two isopropyl groups and six singlet methyls. The 13C and DEPT NMR spectroscopic data showed four noticeable quaternary signals for abietane diterpenoid at δ C 40.1 (s, C-4), δ C 46.7 (s, C-10), δ C 33.8 (s, C-4′), δ C 39.3 (s, C-10′) [19, 20]. These evidences indicated that compound 1 should be a dimer of two abietane diterpenoids units.
Analysis of the 1D and 2D NMR spectra distinguished two sets of diterpenoid signals, C-1–C-20 and C-1′–C-20′, respectively. According to the characteristic signals for normal abietane diterpenoids at δ C 40.1 (s, C-4), δ C 46.7 (s, C-10), δ C 24.3 (q, C-18), δ C 30.8 (q, C-19), δ C 24.2 (q, C-20), and an isopropyl group at δ C 26.9 (d, C-15), δ C 21.5 (q, Me-16), and δ C 21.0 (q, Me-17), the structure of unit 1 can be ascribed to be an abietane diterpenoid [21–23]. The HMBC correlations (Fig. 1) from Me-20 (δ H 1.40) to C-1 (δ C 31.7), C-5 (δ C 80.2), C-9 (δ C 124.7), and C-10; from Me-18 (δ H 1.32) and Me-19 (δ H 1.02) to C-3 (δ C 38.9), C-4, and C-5; from H-15 (δ H 26.9) to C-12 (δ C 181.8), C-13 (δ C 141.3), and C-14 (δ C 135.0); from H-14 (δ H 6.55) to C-7 (δ C 141.2), C-9, and C-12; and from H-6 (δ H 5.42) to C-8 (δ C 140.8), together with proton spin systems H-1/H-2/H-3 and H-6/H-7 obtained from the 1H–1H COSY spectrum (Fig. 1), established the structure of the unit 1.
The other unit was also deduced to be an abietane diterpenoid based on the characteristic quaternary signals at δ C 33.8 (s, C-4′) and δ C 39.3 (s, C-10′), the typical methyls at δ C 33.9, δ C 22.2, δ C 20.4 for Me-18′, Me-19′, and Me-20′, respectively, and the isopropyal group at δ C 27.6 (C-15′), δ C 22.3 (Me-16′), and δ C 21.5 (Me-17′). The planar structure of this unit was elucidated to be almost identical with that of the known analogue, dethdroabietane-11,12-diol [24], based on the comparative analysis of their NMR spectral data and the HMBC correlations from Me-20′ (δ H 1.43) to C-1′ (δ C 36.9), C-5′ (δ C 53.0), C-9′ (δ C 135.0), and C-10′; from Me-18′ (δ H 0.95) and Me-19′ (δ H 0.95) to C-3′, C-4′, and C-5′; from H-15′ (δ H 27.6) to C-12′ (δ C 133.8), C-13′ (δ C 132.8), and C-14′ (δ C 119.5); from H-14′ (δ H 6.36) to C-7′ (δ C 32.4), C-9′, and C-12′; and from H-5′ (δ H 1.26) to C-4′ and C-6′ (δ C 19.3), together with two proton spin systems, H-1′/H-2′/H-3′ and H-6′/H-7′, observed from the 1H–1H COSY spectrum (Fig. 1).
The two units account for 13 degrees of unsaturation. Since the totally degrees of unsaturation were 14, the remained one degree of unsaturation should be ascribed to the linkage between the two units through C-11/C-5/C-6 to C-11′/C-12′ to create an additional ring. The HMBC of H-6/C-12′ confirmed the linkage of C-6/C-12′ through an oxygen atom. In the 1H NMR spectrum of 1, an obvious OH signal can be found at δ H 7.50, and this OH group can be deduced to be attached at C-11 as evidenced by its HMBC correlations with C-9 (δ C 124.7), C-11 (δ C 144.1), and C-12 (δ C 181.8). Then, the remained oxygenated quaternary at (C-5) and the oxygenated aromatic quaternary carbon (C-11′) were deduced to linked through ether bridge.
In the NOESY spectrum (Fig. 1), diagnostic cross-peaks of H-6/Me-20, Me-19/H-6, and Me-19/Me-20 indicated the β-orientation of H-6, Me-19, and Me-20. The NOE correlations of Me-20′/Me-19′ and Me-18′/H-5′ indicated that H-5′ was α-oriented. In addition, the α-substitution of O-5 was suggested by analysis of the molecular model of 1, otherwise the NOE correlation of H-6/Me-19 should be unobservable.. Thus, the structure of 1 was elucidated and named salviwardin A.
The molecular formula of salviwardin B (2) was determined to be C40H52O4 from its 13C NMR and HRESIMS spectral data, indicating one more unsaturation than 1. Comparison of their 1D and 2D NMR data indicated that the structures of 1 and 2 were similar to each other (Tables 1, 2). The difference lied in that the two methylenes (C-6′ δ C 19.3 and C-7′ δ C 32.4) in 1 were replaced by a double bond (C-6′ δ C 126.9 and C-7′ δ C 128.0) in 2, which indicated that 2 was a 6′,7′-dehydrogen derivative of 1. This was confirmed by HMBC correlations from H-6′ (δ H 5.82) to C-4′ (δ C 33.3), C-5′ (δ C 51.6) and C-8′ (δ C 125.8), and from H-7′ (δ H 6.37) to C-5′and C-14′ (δ C 118.0). By detailed analysis of its ROESY (Fig. 2) spectrum, the relative configuration of 2 was also elucidated to be the same as that of 1. Ultimately, the structure of 2 was determined and named salviwardin B.
Salviwardin C (3) was assigned the molecular formula C20H24O2 by analysis of its 13C NMR and HREIMS (m/z 296.1775, [M]+). Comparing of NMR spectroscopic data of 3 (Table 3) with that of naphth-[1,8-bc]-oxocin-11-ol,2,3,4,5tetrhydro-2,2,6-trimethyl-10-(1-methlethyl) indicated that the two compounds are similar to each other. The difference lied in that the C-3, C-4, and Me-18 in the known compound were replaced by an oxygenated methine at δ C 86.5 (C-3) and a part of double-bond (C-4, δ C 145.5 and C-18, δ C 111.0) in 3 [25]. The HMBC correlations from H-3 (δ H 4.50) to C-1 (δ C 26.0), C-2 (δ C 33.6), C-4 (δ C 145.5), C-11 (δ C 141.0), C-18 (δ C 111.0) and C-19 (δ C 18.8), and from H-18 (δ H 5.10 and 4.90) to C-3 (δ C 86.5), C-4 (δ C 145.5), C-19, together with the proton spin systems H-1/H-2/H-3, determined the structure of A ring of 3 as shown in Fig. 3. Other parts of 3 were identical to those of the known compound by detailed analysis of the 1H–1H COSY and HMBC correlations (Fig. 3). Therefore, the structure of 3 was determined and named salviwardin C.
All isolates were tested for their in vitro inhibitory activities against HL-60, SMMC-7721, A549, MCF-7, and SW480 human tumor cell lines using the MTT method described previously [26]. The results indicated that all the compounds were inactive with IC50 > 30 μM.
3 Experiment Section
3.1 General Experimental Procedures
Optical rotations were obtained with a Jasco P-1020 polarimeter. UV spectra were measured on Shimadzu UV-2401A spectraphotometer. IR spectra were detected on a Bruker Tensor-27 infrared spectrophotometer with KBr pellets. 1D and 2D NMR spectra were recorded on Bruker AV-400, and Avance III-600 MHz spectrometers with TMS as the internal standard. Chemical shifts (δ) were expressed in ppm with reference to the solvent signals. HRESIMS analysis and HREIMS were determined on API QSTAR time-of-flight spectrometer and on Waters Auto spec Premier P776 mass spectrometer. Semi-preparative HPLC was performed on an Agilent 1100 liquid chromatography with a Zorbax SB-C18 (9.4 mm × 25 cm) column. Column chromatography was performed on Sephadex LH-20 (GE Healthcare), Silica gel (100–200 and 200–300 mesh, Qingdao Marine Chemical Co., Ltd., Qingdao, China), and Amphichroic RP-18 gel (40–63 μm, Merck, Darmstadt, Germany) and MCI gel (75–150 μm, Mitsubishi Chemical Corporation, Tokyo, Japan). Fractions were monitored by TLC and spots were visualized by heating silica gel plates sprayed with 10 % H2SO4 in EtOH.
3.2 Plant Material
The roots parts of S. wardii were collected in Zuogong prefecture Tibet, China, in July 2011. The plant was identified by Dr. Yu-Kun Wei, Shanghai Chenshan Plant Science Research Center, Chinese Academy of Sciences. A voucher specimen was deposited in Kunming Institute of Botany,Chinese Academy of Sciences with identification number 20110712.
3.3 Extraction and Isolation
The roots parts of the air-dried S. wardii (33 kg) were powdered and percolated with acetone at room temperature and filtered. The filtrate was evaporated in vacuo to be concentrated. The crude extract (1.6 kg) was subjected to silica gel column chromatography eluted with CHCl3, EtoAc and MeOH, respectively. The fraction CHCl3 with petroleum-CHCl3-EtoAc gradient (50:1:1, 20:1:1, 10:1:1, and 5:1:1) to produce seven fractions, A–G. Fraction B (223 g) was separated over a MCI gel column (MeOH-H2O from 70:30 to 100:0) to obtain eight fractions (Fr. B1–B8). Fr. B1 (12 g) was isolated over an RP-18 gel column (MeOH–H2O from 85:15 to 100:0) to obtain seven fractions (Fr. B2a–B2g). Fr. B2a (2 g) was separated on a silica gel column, eluted with petroleum ether-acetone (from 100:1 to 50:1), to yield six fractions (B2a1–B2a6). Fr. B2a2 was purified by repeated silica gel columns and semipreparative HPLC (RP-18, 98 % MeCN-H2O) and TLC to afford 1 (80 mg), 2 (13 mg). Fr. B3 (27 g) was separated over a MCI-gel column (MeOH-H2O from 85:15 to 100:0) to obtain six fractions (Fr. B3a–B3f). Fr. B3e (5 g) was then chromatographed on a silica gel column, eluted with petroleum ether-acetone (from 50:1 to 5:1), to yield eight fractions (Fr. B3e1–B3e8). Subfraction B3e1 (110 mg) was chromatographed by semipreparative HPLC (89 % MeOH-H2O) to afford two fractions (Fr. B3e1a–Fr. B3e1b). Fr. B3e1a (15 mg) was chromatographed by semipreparative HPLC (92 % MeCN-H2O) to afford 3 (10 mg). Subfraction B3e1 (1.7 g) was chromatographed by semipreparative HPLC (83 % MeOH-H2O) to afford two fractions (Fr. B3e2a–Fr. B3e1f). Fr. B3e1a (15 mg) was chromatographed by semipreparative HPLC (92 % MeCN-H2O) and chromatographed on a silica gel column, eluted with petroleum ether-acetone (100:1) to afford prionitin (4, 100 mg). Fr. B3f (3.3 g) was then chromatographed on a silica gel column, eluted with petroleum ether-acetone (from 50:1 to 5:1), to yield eight fractions (Fr. B3f1–B3f7). Fr. B3f1 (726 mg) was then chromatographed on a silica gel column, eluted with petroleum ether-acetone (50:1), to yield eight fractions (Fr. B3f1a–B3f1f). Fr. Bf1a, Fr. Bf1c, and Fr. Bf1f were purified by semipreparative TLC and chromatographed by semipreparative HPLC (92 % MeCN-H2O) to yield sahandol (5, 49 mg), salvilenone (6, 10 mg), microstegiol (7, 6 mg), and ferruginol (8, 120 mg).
3.4 Salviwardin A (1)
Orange powder; \([\alpha]_{\text{D}}^{16}\) + 325 (c 0.17, CDCl3); UV (CDCl3) λ max (log ε) 374.5 (3.68) nm; IR (KBr) ν max 3441, 2939, 1624, 1476, 1417, 1364, 1296, 1134, 1121, 1011 cm−1; 1H and 13C NMR data, see Tables 1 and 2; positive ESIMS m/z 599 [M + H]+; positive HREIMS m/z 598.4029 (calcd for C40H54O4 [M]+, 598.4022).
3.5 Salviwardin B (2)
Orange powder; \( [\alpha ]_{\text{D}}^{20} \) + 184 (c 0.19, MeOH); UV (MeOH) λ max (log ε) 476 (3.25) nm; IR (KBr) ν max 3442, 2927, 2870, 1626, 1579, 1467, 1394, 1291, 1172, 1088, 1008 cm−1; 1H and 13C NMR data, see Tables 1 and 2; positive ESIMS m/z 619 [M + Na]+; positive HRESIMS m/z 597.3941 (calcd for C40H53O4 [M + H]+, 597.3944).
3.6 Salviwardin C (3)
Colorless oil; \( [\alpha ]_{\text{D}}^{26} \) −2 (c 0.26, CDCl3); UV (CDCl3) λ max (log ε) 336.5 (3.35) nm; IR (KBr) ν max 3430, 2927, 2871, 1722, 1633, 1423, 1330, 1271, 1172, 1023 cm−1; 1H and 13C NMR data see Table 3; positive EIMS m/z 296 ([M]+, 77), 268 (26), 267 (94), 265 (100), 255 (18), 241 (35), 213 (54); positive HREIMS m/z 296.1775 (calcd for C20H24O2 [M]+, 296.1776).
3.7 Cytotoxicity Assays
The following human tumor cell lines were used: HL-60, SMMC-7721, A-549, MCF-7, and SW-480, which were obtained from ATCC (Manassas, VA, USA). All cells were cultured in RPMI-1640 or DMEM medium (Hyclone, Logan, UT, USA), supplemented with 10 % fetal bovine serum (FBS, Hyclone) at 37 °C in a humidified atmosphere with 5 % CO2. Cell viability was assessed by conducting colorimetric measurements of the amount of insoluble formazan formed in living cells based on the reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma, St. Louis, MO, USA). Briefly, 100 μL of adherent cells was seeded into each well of a 96-well cell culture plate and allowed to adhere for 12 h before test compound addition, while suspended cells were seeded just before test compound addition, both with an initial density of 1 × 105 cells/mL in 100 μL of medium. Each tumor cell line was exposed to the test compound at various concentrations in triplicate for 48 h, with cis-platin and paclitaxel (Sigma) as positive control. After the incubation, MTT (100 μg) was added to each well, and the incubation continued for 4 h at 37 °C. The cells were lysed with 100 μL of 20 % SDS −50 % DMF after removal of 100 μL of medium. The optical density of the lysate was measured at 595 nm in a 96-well microtiter plate reader (Bio-Rad 680). The IC50 value of each compound was calculated by Reed and Muench’s method [25].
References
H.M. Cheng, K.P. Cheng, T.F. Chang, H.F. Chow, K.Y. Chui, P.M. Hon, F.W. Lau, Y. Yang, Z.P. Zhong, J. Org. Chem. 55, 3537–3543 (1990)
J.M. Chang, N.S. Du, Chin. Tradit. Pat. Med. 21, 480–482 (1999)
L. Rodriguez-Hahn, B. Esquivel, A.A. Sanchez, J. Cardenas, O.G. Tovar, M. Soriano-Garcia, A. Toscano, J. Org. Chem. 53, 3933–3936 (1988)
L. Rodriguez-Hahn, R. O’Reilly, B. Esquivel, E. Maldonado, A. Ortega, J. Cardenas, R.A. Toscano, J. Org. Chem. 55, 3522–3525 (1990)
M. Xue, Y.B. Shi, Y. Chui, B. Zhang, Y.J. Luo, Z.T. Zhou, W.J. Xia, R.C. Zhao, H.Q. Wang, Nat. Prod. Res. Dev. 12, 27–32 (2000)
L.H. Meng, J.S. Zhang, J. Ding, J. Biochem. Pharm 62, 733–741 (2001)
X.H. Wang, K.F. Bastow, C.M. Sun, Y.L. Lin, H.J. Yu, M.J. Don, T.S. Wu, S. Nakamura, K.H. Lee, J. Med. Chem. 47, 5816–5819 (2004)
A. Munro, M.A. Rizzacasa, B.L. Roth, B.A. Toth, F. Yan, J. Med Chem. 48, 345–348 (2005)
R.S. Xu, Danshen. Academic Press, Beijing. p.81 and 158 (1990)
T. Tanake, A. Nishimu, I. Kouno, G.I. Nonake, T.J. Young, J. Nat. Prod. 59, 843–849 (1996)
W.S. Chen, X.M. Jia, W.D. Zhang, Z.Y. Lou, C.Z. Qiao, Acta Pharm. Sin. 38, 354–357 (2003)
X.R. Luo, Guangdong Press of Science & Technology. 3, 37 (1994)
Kunming Institute of Botany, Chinese Academy of Sciences, Flora of China, vol. 66 (Science Press, Beijing, 1977), pp. 83–84
A.R. Jassbi, M. Mehrdad, F. Eghtesadi, S.N. Ebrahimi, I.T. Baldwin, Chem. Biodivers. 3, 916–922 (2006)
S.N. Ebrahimi, S. Zimmermann, J. Zaugg, M. Smiesko, R. Brum, M. Hamburger, Plant Med. 79, 150–156 (2003)
K. Takenori, O. Takashi, H. Teruo, K. Hiroshi, Phytochemistry 24, 2118–2120 (1985)
R.A. Taj, J.R. Green, J. Org. Chem. 75, 8258–8270 (2010)
J.J. Gao, G.Q. Han, Phytochemistry 44, 759–761 (1997)
Y.H. Kuo, M.T. Yu, Chem. Pharm. Bull. 44, 1431–1435 (1996)
B. Li, F.D. Niu, Z.W. Zhang, D.Z. Wang, H.D. Sun, Phytochemistry 30, 3815–3817 (1991)
S. Hasegawa, Y. Hirose, Phytochemistry 21, 643 (1982)
M. Tada, T. Hara, C. Hara, K. Chiba, Phytochemistry 45, 1475 (1997)
A. Ulubelen, S. Oksuz, G. Topca, A.C. Goren, W. Voelter, J. Nat. Prod. 64, 549 (2001)
A.J. Sanchez, J.P. Konopelski, J. Org. Chem. 59, 5445–5452 (1994)
F. Conti, C.H. Eugster, W. Von Philipsborn, Helv. Chim. Acta 49, 2267–2274 (1996)
M.C. Alley, D.A. Scudiero, A. Monks, M.L. Hursey, M.J. Czerwinski, D.L. Fine, B.J. Abbott, J.G. Mayo, R.H. Shoemaker, M.R. Boyd, Cancer Res. 48, 589–601 (1988)
Acknowledgments
The work was financially supported by the foundations from NSFC (81373291), the National Science and Technology Support Program of China (2013BAI11B02), the Young Academic Leader Raising Foundation of Yunnan Province (No. 2009CI073), and the foundation from CAS to Dr. G. Xu.
Conflict of Interest
All authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Xiao, QL., Xia, F., Yang, XW. et al. New Dimeric and seco-Abietane Diterpenoids from Salvia wardii . Nat. Prod. Bioprospect. 5, 77–82 (2015). https://doi.org/10.1007/s13659-015-0054-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13659-015-0054-6



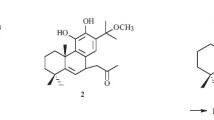

 ), 1H-1H COSY (
), 1H-1H COSY ( ), and ROESY (
), and ROESY ( ) correlations of 1
) correlations of 1

