Abstract
Pancreatic and colorectal cancer are two of the most lethal cancers; this stems from poor prognosis. Current treatments may lack effectiveness and produce dangerous side effects, hence a need for alternatives. Medicinal plants like Millettia griffoniana may possess anti-cancer properties. This study aimed to isolate and identify compounds from M. griffoniana to test for potential anti-cancer activity on pancreatic and colorectal cancer. Seeds and root bark of M. griffoniana underwent solvent extraction and separation using column chromatography. Isolated compounds were screened using AlamarBlue assays and cytotoxic compounds were identified with 2D NMR. Compounds were subject to cellular viability assays using AlamarBlue and xCELLigence analysis. The Caspase Glo®-3/7 kit measured caspase activity and Real-Time PCR analysis measured apoptosis-related gene expression. 7 compounds were isolated and screened. Compounds 5 and 7 were chosen; identified as durmillone and isojamaicin. Both showed varying concentration-dependent cytotoxic activity, in AlamarBlue and xCELLigence assays, for both cell lines. Caspases 3 and 7 were up-regulated and both compounds up-regulated BAX and down-regulated BCL-2 and p53 in both cell lines. Durmillone and isojamaicin displayed cytotoxic activity on pancreatic and colorectal cancer. Apoptotic activity induced by the compounds was verified by the up-regulation of caspase activity and BAX and down-regulation of BCL-2 and p53. Further studies need to be done towards understanding the mechanisms by which these compounds bring about their cytotoxic activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer, in essence, is a disease caused by the build-up of alterations or mutations in the natural genome of somatic body cells, resulting in abnormal proliferation and cell growth (DeVita et al. 2015). Cancer and tumour formation is driven by a multistep process reflecting alterations in the genome (Hanahan and Weinberg 2011). Cancerous cells bypass cellular regulatory pathways that control cell homeostasis, proliferation and cell death allowing them to acquire distinct abilities. Such abilities include self-regulating growth signals, the incapability to respond to growth-inhibitory (antigrowth) signals, evasion of apoptosis, the ability to replicate infinitely, continuous angiogenesis, and the ability to invade other tissues. Cells that acquire these abilities have proven that they have breached anti-cancer mechanisms instilled by the somatic cells, which ultimately leads to the formation of tumours (Hanahan and Weinberg 2011). Pancreatic cancer is the 11th most commonly diagnosed cancer, based on statistics provided by Globacon 2020 (Sung et al. 2020), and there has been a drastic increase in the incidence rate of pancreatic cancer and is expected to increase by an estimated 77% by the year 2040 (Rawla et al. 2019). Of the 496 000 cases reported in 2020, 466 000 of those cases resulted in deaths, making it the 7th most lethal cancer. The lethality of this cancer stems from the poor prognosis of pancreatic cancers as patients have been found to experience symptoms in the later stages of cancer development (Mizrahi et al. 2020). A similar trend is seen for colorectal cancers where symptoms typically appear in the later stages of development when the cancer has become aggressive, malignant and is usually in the metastatic stages, and numerous other organs may have possibly been affected (Xi and Xu 2021). This accounts for colorectal cancer being the third most commonly diagnosed cancer and the second most fatal; as 9.4% of the globally reported cancer cases (865 630 reported cases) and 10.6% of cancer-related deaths (1 065 930 deaths) in 2020 were related to colorectal cancer (Sung et al. 2020).
Numerous treatments are available to combat cancer, however, excessive costs, ineffective treatments and harsh side effects drive the need to find alternative forms of cancer treatment and the development of more effective drugs. Medicinal plants such as Millettia griffoniana have been used in numerous traditional healing practices. They have been proven to display a range of biological activities such as bactericidal, anti-fungal, anti-cancer and anti-inflammatory properties (Banzouzi et al. 2008). The established medical benefits of this plant make it a potential candidate to identify the chemical and pharmacological components of M. griffoniana for the sake of finding anti-cancer active components which can be used for future anti-cancer drug development. In this study, compounds were isolated from extracts of M. griffoniana and screened for potential anti-cancer activity. Compounds showing potential were tested further on pancreatic and colorectal cancer.
Materials and methods
Isolation process
The compounds, to be tested on the relevant cancer cells, were obtained from the seeds of and root bark of M. griffoniana as described by Ngamga et al. (2005) which were obtained from the Cameroon National Herbarium and a voucher specimen documenting the collection was deposited under the number 32315/SRF/HNC. A mass of 2 kg of seeds was dried and extracted for 24 h, using 60 L of acetone (University of Johannesburg Chemical Store). The dried extract was then separated using vacuum Silica (Merck, Germany) based column chromatography using a gradient-based mobile phase of ethyl acetate (EtOAc) (University of Johannesburg Chemical Store) and petrol (University of Johannesburg Chemical Store). This extract yielded 130 × 500 ml fractions which were analysed and combined using TLC resulting in 4 different fractions; namely fraction A (9 petrol:1 EtOAc), series B (4 petrol:1 EtOAc), series C (1 petrol:1 EtOAc) and series D (100% EtOAc). The 15 g obtained as series C had undergone further purification using a Sephadex LH 20 (Sigma Aldrich, Germany) in methanol (University of Johannesburg Chemical Store). The root bark of M. griffoniana (50 g of the sample obtained from the same source) had undergone extraction using chloroform (University of Johannesburg Chemical Store) as a solvent, following the same procedure as described previously for the powdered seeds. The dried extract had been subjected to silica-based flash column chromatography using petrol and EtOAc.
Cell culture
The HEK 293 (obtained from Sibusiso Malindisa, University of South Africa), MIA PaCa 2 and HT 29 cells (both obtained from Prof. Raymond Motadi, University of Johannesburg) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Sigma Aldrich, USA) supplemented with 10% foetal bovine serum (FBS) (Biogen, UK) and 1% Pen-Strep (penicillin–streptomycin) (Biowhitakker, Germany) at 37 °C in 95% humidity and 5% CO2.
Compound preparation
The compounds extracted from M. griffoniana were made up to a stock concentration of 100 mg/ml by weighing them out and dissolving them in the appropriate amount of 100% dimethyl sulphoxide (DMSO) (PanReac AppliChem, Germany) (based on the weight). The compounds were stored at 4 °C until ready to use.
AlamarBlue cellular viability assay: screening
The HEK 293, MIA PaCa 2 and HT 29 cells were plated in 96 well plates (at 1 × 106 cells per plate. Column 1 of the plate contained 100 µl of media. Column 2 was left untreated and columns 2–3 were treated with the negative 0.1% DMSO and the positive control 100 µM etoposide (Sigma Aldrich, Germany). Columns 5–11 were treated with the 7 chosen compounds made up to 100 µl/ml. The AlamarBlue reagent was added to each well following the 24 h treatment. This was done by adding 10 µl of AlamarBlue (Thermofisher, USA) before incubating the plate for 2 h at 37 °C. The fluorescence was measured in each well using a plate reader and the Gen 5 programme 530/25 nm excitation and 590/35 nm emission.
Nuclear magnetic resonance (NMR) and structural elucidation
The extracted compound chosen to be studied further (following the screening process) was analysed using 2D NMR (1H–1H, 13C–13C, DEPT, COSY, HMQC, HMBC and NOESY) to determine the structure of the compound. The identity of the compound was obtained by comparing the 1H and 13C spectra and verifying the compound identity based on an IUPAC name and referral to literature.
AlamarBlue cellular viability assay: cytotoxicity test for chosen compounds
To test the cytotoxicity of the compounds chosen for further studies, the compounds had undergone a concentration-based study to find a general pattern of cytotoxicity and an IC50 for the treatments. The cells were plated and treated with the controls in the same way as the screening process. For columns 5–10, which contained varying concentrations of the compound treatment, diluted serially 2X from 100 to 3.125 µg/ml of the compound. The fluorescence reading for the plate was read in the same way as the screening process. An IC50 was calculated based on the % cellular viabilities for the treated cells.
xCELLigence: real-time analysis of cellular viability
An xCELLigence DP system was used to conduct a real-time analysis of cellular viability changes brought about by the compounds of interest. The programme was set us per the manufacturer’s protocol. The experiment was calibrated to accommodate for a media blank, to calculate the cell index without the influence of the medium, by adding 80 µl of media and reading it on the instrument for one cycle. The media was discarded and then the appropriate cells (from the MIA PaCa 2 HT 29 and HEK 293 cell lines) were added to the wells of the xCELLigence 16-well E-plate (except for the first set of duplicate wells which only contained media) to a volume of 5000cells/100 µl. the cell growth was monitored for 24 h. Following the 24 h growth period, the cells were then treated in the same way as the AlamarBlue experiment (negative control 0.1% DMSO, positive control 100 µM etoposide and the compound treatment 100–12.5 µg/ml). The cells were monitored for 48 h following the treatment.
Caspase Glo® 3/7 activity assay
The MIA PaCa 2 and HT 29 cell lines were subjected to a caspase detection assay to determine if caspase 3 and 7 were up-regulated due to the compound treatment. This was done using the Caspase Glo® 3/7 assay kit (Promega, USA) and conducted based on the manufacturer’s protocol.
Real-time polymerase chain reaction (PCR)
RNA extraction
RNA was extracted for the untreated cells as well as cells treated with the IC50s of the compounds on the MIA PaCa 2 and HT 29 cell lines. The cells were plated and treated in the same manner as the caspase assay and microscopy. RNA extraction was done using the ReliaPrep™ RNA Cell Miniprep System (Promega, USA) and conducted according to the kit protocol. The quality and quantity of the extracted RNA were checked using a nanodrop and purified RNA was stored at –70 °C until needed.
cDNA synthesis
The extracted RNA had undergone reverse transcription using the GoScript™ Reverse Transcription System (Promega, USA). The cDNA was synthesised by combining 2 µl of the extracted RNA with 2 µl of the Random Primer and 1 µl nuclease-free water which was then heated at 70 °C for 5 min. The RNA samples were chilled on ice for 5 min and a 15 µl transcription mix was made up by combining the components of the GoScript™ Reverse Transcription System according to the manufacture’s protocol as given below:
Volume (µl) | Component |
|---|---|
4 | GoScript™ 5X reaction buffer |
2 | MgCl2 |
1 | GoScript™ reverse transcriptase |
8 | Nuclease-free water |
The RNA sample combined with the transcription mix was synthesised into cDNA using a thermocycler with conditions set per the GoScript™ Reverse Transcription System protocol as given below:
Thermocycler condition | Temperature (°C) | Time |
|---|---|---|
Annealing | 25 | 5 min |
Extension | 42 | 1 h |
Deactivation of reverse transcriptase enzyme | 70 | 15 min |
Hold | 4 | 5 min |
Real-time PCR
The cDNA samples were prepared for Real-time PCR using the iTaq™ Universal SYBR® Green Supermix (Biorad, USA) as per the manufacturer’s protocol given below:
Volume (µl) | Component |
|---|---|
5 | iTaq Universal SYBR Green Supermix |
1 | Primers |
1 | cDNA template |
3 | Nuclease-free water |
The cDNA samples were analysed using the Biorad CFX96-connect machine and analysis software which was set up as given below:
Thermocycler condition | Temperature | Time |
|---|---|---|
Initial denaturation | 95 °C | 5 min |
Second denaturation | 95 °C | 15 s |
Annealing | 55 °C | 30 s |
Extension | 72 °C | 30 s |
Number of cycles | 40 | |
Final extension | 72 °C | 5 min |
Melt curve analysis | 65–95 °C | 2 s |
Statistical analysis
Statistical analysis of the results done in triplicate was conducted using the Graph Pad Prism software to average results and to conduct a T-test between the results presented by the treated samples in comparison to the untreated samples. This was used to indicate if any significant difference was shown between the results using asterisks (*) expressed as (*P < 0.05, **P < 0.01 ***P < 0.001) where (P < 0.05) which was considered as significant.
Results
Compound isolation and screening using AlamarBlue cellular viability assay
Following the further purification of Series C from the acetone crude extract of M. griffoniana, using Sephadex LH-20, as well as the flash chromatography of the chloroform extract, yielded 7 compounds. The isolated compounds were labelled as numbers 1–7 as indicated in Table 1 below:
The durmillone, odorantine and MTF 51 are compounds which had already undergone 2D NMR and structural elucidation for other studies (conducted by Jordan Tonga Lembe, University of Johannesburg Department of Chemistry, and Tagatsing Fotsing Maurice and Yankep Emmanuel, University of Yaoundé) and the compounds identified with the name griffoniana have not and hence been given a number to indicate which fraction they were found in the elution series. The purity of the compounds was checked using proton NMR. Structural elucidation using 2D NMR was only done for the compounds chosen for further studies.
Compounds 1–7 were tested against each other at a constant concentration of 100 µg/ml for two different treatment times on the MIA PaCa 2 cell line with a positive control set as 100 µM etoposide and a negative control set as 0.1% DMSO. Based on the resulting % viabilities and the calculated % cytotoxicities from Fig. 1A and B, compounds 5 (durmillone) and 7(griffoniana 55) were picked for further studies.
The average percentage cellular viability readings for the 24 h (A) and 48 h (B) treatment of MIA PaCa 2 showing the untreated cells, the cells treated by 0.1% DMSO, 100 µM etoposide, and the compounds extracted from M. griffoniana (100 µg/ml). The asterisk (*) represents (*P < 0.05, **P < 0.01 ***P < 0.001, ****P < 0.0001) as calculated by a T-test between untreated and treated samples
NMR and structural elucidation of compounds chosen for further studies
NMR was used to obtain the structure and identity of griffoniana 55 (as durmillone had been identified previously and a structure was obtained). Griffoniana 55 was identified as Isojamaicin (3ʹ-Methoxy-4ʹ, 5ʹ-methylenedioxy-6ʺ, 6ʺdimethylpyrano [2ʺ,3ʺ:7,8] isoflavone) a white amorphous powder that was soluble in chloroform. Its molecular formula was established to be C22H18O6 from HREIMS, displaying a molecular ion at m/z = 378.1204 which was done in a study conducted by Ngamga et al. ( (Ngamga et al. 2005)). Durmillone and isojamaicin share the same molecular formulae and the same structure (Fig. 2A and B). Tables 1 and 2 show a comparison between the positions of peaks in the 1H and 13C spectra for durmillone and isojamaicin. The only difference between durmillone and isojamaicin is the presence of the methoxy group in isojamaicin present on C3̍ and 4̍ of ring B (as shown in Fig. 2A) (Table 3).
Cytotoxicity test on chosen compounds of interest isolated from M. griffoniana using AlamarBlue cellular viability assay
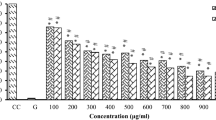
Following the identification of durmillone and isojamaicin, both compounds were subject to a concentration-series-based AlamarBlue assay on MIA PaCa 2, HT 29 and HEK 293, to examine the potency of their activity and generate an IC50 for further studies. For the MIA PaCa 2 cell line (Fig. 3A) durmillone was observed to show a concentration-dependent effect with an IC50 of 104.69 µg/ml. For the MIA PaCa 2 cells treated with isojamaicin (Fig. 3B), isojamaicin is shown to display a concentration dependant form of cytotoxic activity, with an IC50 of 26.45 µg/ml.
The average percentage cellular viability readings for the 24 h treatment with the compounds of interest (treated at 100, 50, 25, 12.5, 6.25 and 3.125 µg/ml). The MIA PaCa 2 cell line is shown as A (durmillone treated) and B (isojamaicin treated), the HT 29 cell line is shown as C (durmillone treated) and D (isojamaicin treated) and the HEK 293 cell line is shown as E (durmillone treated) and F (isojamaicin treated). The asterisk (*) represents (*P < 0.05, **P < 0.01 ***P < 0.001, ****P < 0.0001) as calculated by a T-test between untreated and treated samples
When tested on the HT 29 cell line (Fig. 3C), durmillone, once, again, showed a concentration-based pattern of cytotoxic activity. The IC50 of 79.22 µg/ml for the HT 29 cells line is significantly lower than the IC50 on the MIA PaCa 2 cell line suggesting that the cytotoxic effects of durmillone are potentially influenced by the cell line it is used on. the cytotoxic effects of the HT 29 cell line seem to be less prominent than the effect seen of the MIA PaCa 2 cell line when treated with isojamaicin (Fig. 3D). This can also be seen by the difference in IC50 values which was calculated as 372.6 µg/ml for the HT 29 cell line.
For the durmillone treated HEK 293 cells, (Fig. 3E), mild cytotoxicity is found at 100, 50 and 6.25 µg/ml and the IC50 for durmillone was calculated as 1065.4 µg/ml, which is significantly greater than the tested concentrations. For isojamaicin, (Fig. 3F), mild levels of cytotoxicity are seen in the 100, 50 12.5 and 6.25 µg/ml treatments and the IC50 was calculated as 2521.8 µg/ml, of which, much like the durmillone, is significantly greater than the tested concentrations.
xCELLigence real-time cellular viability analysis
Following the 24 h cell proliferation time period, the cells were treated with the same controls and concentrations of durmillone and isojamaicin (in a serially diluted concentration series from 100 to 12.5 µg/ml based on how many wells were available). Figure 4A shows the durmillone treated MIA PaCa 2 cells. A lowering of the cell index readings indicated cell death. The highest cell index reading is shown for the untreated, DMSO treated and 12.5–25 µg/ml durmillone treated cells and the 50–100 µg/ml durmillone treatments showed lower cell index readings compared to the 12.5–25 µg/ml. Figure 4B depicts the isojamaicin treated MIA PaCa 2 cells The lowest cell index readings can be seen in the etoposide and 100 µg/ml isojamaicin treated cells. The pattern of cytotoxic activity shows signs of concentration dependence where the cell index increased slightly as the concentration of isojamaicin decreased, however, a comparison is not as distinct as the durmillone treatments.
xCELLigence real-time cellular viability analysis of durmillone and isojamaicin treated cells showing the cell index given every 15 min for a time period of 72 h (24 h cell proliferation and 48 h treatment monitoring) the graph depicts the media (red), the untreated cells (light green), 0.1% DMSO (dark blue) and 100 µM Etoposide (pink) and the durmillone or isojamaicin treatment as 100 µg/ml (light blue), 50 µg/ml (purple), 25 µg/ml (brown) and 12.5 µg/ml (dark green). The MIA PaCa 2 cell line is shown as A (durmillone treated) and B (isojamaicin treated), the HT 29 cell line is shown as C (durmillone treated) and D (isojamaicin treated) and the HEK 293 cell line is shown as E (durmillone treated) and F (isojamaicin treated)
With regards to the HT 29 cell line, Fig. 4C shows the durmillone treated HT 29 cells with the lowest cell index readings were shown by the etoposide and 100 µg/ml durmillone treated cells, indicating a higher level of cell death (which much like what was seen in the MIA PaCa 2 cell line in Fig. 4A and D depicts the isojamaicin treatment on the HT 29 cell line. The lowest cell index readings were shown by the isojamaicin treated cells, indicating a higher level of cell death and the lowest cell index readings were shown by the untreated and DMSO treated cells, indicating lower levels of cell death.
In the same manner and conditions as the AlamarBlue assay, durmillone and isojamaicin were tested on HEK 293 to determine their relative cytotoxic activity on non-cancerous human cell lines. Figure 4E depicts the HEK 293 cell line, following the treatment with durmillone, which was shown to have low and insignificant levels of cytotoxicity in a relatively concentration-dependent manner. Mild levels of cytotoxic activity are seen in Fig. 4E as indicated by lower cell index readings (when compared to the untreated cells), however, the decrease in cell index readings was little and the readings eventually began to increase and cluster with the readings for the DMSO treated and the untreated cells. A sign of concentration-based cytotoxicity is evident as the 100 and 50 µg/ml durmillone treatments generally had lower cell index readings compared to the 25 and 12.5 µg/ml treatments. Figure 4F shows the isojamaicin treated HEK 293 cells of which a higher level of cytotoxic activity brought about by isojamaicin is seen by the lower recorded cell index readings given by the isojamaicin treated cells, however, there are indications of the extent of the cytotoxic activity not being as prominent, meaning the compound is not significantly toxic on HEK 293. When compared to etoposide, etoposide is the only treatment that showed a significant decrease in cell index readings. The isojamaicin treated cells also showed the same pattern of changes in cell index readings as the untreated cells. The cell index readings for the isojamaicin treated cells are also fairly similar to that of the DMSO treated cells (where minimal or no cytotoxic activity is expected), indicating lower levels of cytotoxic activity brought about by the isojamaicin treatment.
The IC50s generated by the xCELLigence RTCA programme were 32.9 µg/ml durmillone and 55.46 µg/ml isojamaicin for the PACA 2 cell line and 25.33 µg/ml durmillone and 39.53 µg/ml isojamaicin for the HT 29 cell line respectively. This differs in comparison to the IC50s generated from the AlamarBlue assays. the IC50s generated by the xCELLigence programme differed at each time point and an appropriate calculated IC50 was chosen based on, firstly, which periods of time showed optimum activity (which was not necessarily 24 h after treatment like what was done for AlamarBlue) and secondly, where the regression squared value was the highest (closest to 1).
Durmillone displayed low levels of cytotoxic activity on HEK 293 and thus presented an IC50 of 2710 µg/ml which is significantly higher than the tested concentration range. An IC50 of 85.08 µg/ml was calculated using the RTCA programme for isojamaicin on the HEK 293 cells. These IC50s are significantly higher than the IC50s calculated for the MIA PaCa 2 and HT 29 cells, showing less cytotoxic activity on the HEK 293 cell line.
Caspase 3 and 7 activity detection
The caspase assay on the MIA PaCa 2 cell line shows evident increase in caspase activity following their treatment with the IC50s of durmillone and isojamaicin (Fig. 5A). The durmillone treated cells showed a 185.37% increase in caspase activity, from 790.3RLU for the untreated cells to 1465RLU for the durmillone treated cells. The isojamaicin treated cells showed an increase of 137.8% (to 1089.3RLU). The caspase assay on the HT 29 cell line was also shown to display an evident increase in caspase activity following their treatment with the IC50s of durmillone and isojamaicin (Fig. 5B). The durmillone treated cells showed a 336.84% increase in caspase activity, from 1647.3RLU for the untreated cells to 5548.7RLU for the durmillone treated cells. The isojamaicin treated cells showed an increase of 263.60% (to 4342.3RLU).
The average luminescence readings (in relative light units) for the 24 h treatment of the cancer cell lines with the compounds of interest and showing the luminescence readings for the untreated cells, the cells treated with 0.1% DMSO, 100 µM etoposide and the IC50s of durmillone (D) and isojamaicin (I) The MIA PaCa 2 cell line is depicted in A with the IC50s for durmillone as 32.9 µg/ml and isojamaicin as 55.46 µg/ml. the HT 29 cell line is depicted in B with the IC50s for durmillone as 25.33 µg/ml and isojamaicin as 39.53 µg/ml. The asterisk (*) represents (*P < 0.05, **P < 0.01 ***P < 0.001, ****P < 0.0001) as calculated by a T-test between untreated and treated samples
Real-time PCR
The down-regulation of the anti-apoptotic gene BCL-2 and the up- regulation of the pro-apoptotic gene, BAX were depicted in Fig. 6. This is seen in both cell lines, for both durmillone and isojamaicin treatments. Durmillone and isojamaicin displayed varying levels of BAX and BCL-2 expression, which ties in with their varying levels of cytotoxic activity depicted in Figs. 3 and 4. Figure 6 depicts the down-regulation of p53 in both the MIA PaCa 2 and HT 29 cell lines after being treated with durmillone and isojamaicin.
The fold expression 2^-ΔΔCt (A and C) and log2(2^-ΔΔCt) (B and D) of apoptosis genes BAX, BCL-2 and p53 against the GAPDH housekeeping gene on the MIA PaCa 2 (A and B) and HT 29 cell line (C and D) shown for untreated cells and the IC50 treated cells for durmillone and isojamaicin for each respective cell line
Discussion
Medicinal plants provide a promising industry for the discovery of new compounds and secondary metabolites that plants produce for their defense mechanism against any attack. In this study we focused on the plant called M. griffoniav which is a known and commonly used medicinal plant in South Africa. Cell viability assay is one important tool at the initial stages of drug discovery this technique involves analysis of the effect of compounds or molecule on a singular cell and measure the cells metabolic processes by which a conclusion is drawn to say the cell is dividing or dead. The results from screening the pure compounds isolated from M. griffoniana depicted that durmillone displayed 51.2% cytotoxicity (where % cytotoxicity = 100 − %cellualr viability) at 24 h, which was the highest % cytotoxicity for that treatment period and at 48 h, the % cytotoxicity decreased to 36.5%. Griffoniana 55 (later identified as isojamaicin) displayed 38.2% cytotoxicity at 24 h which was the third highest % cytotoxicity however at 48 h, it displayed 51.9% cytotoxicity, which was the highest % cytotoxicity for that treatment period. The higher levels of cytotoxicity as well as the ability to remain significantly cytotoxic at two different treatment times made durmillone and griffoniana 55 suitable options for further testing.
Durmillone and isojamaicin are isoflavonoid compounds found abundantly in plants of the Millettia and Lonchocarpus genus. Durmillone and isojamaicin have a characteristic dimethyl pyran group, it is connected to its C6 and C7 of ring A (Yan et al. 2019). Kumar et al. (2017) reviewed several studies showing the anti-cancer activity of pyran-based molecules and their derivatives. Studies like this suggest that cytotoxicity on cancer cells treated by durmillone or isojamaicin may arise from the presence of a pyran group; however, further studies need to be done to verify this observation.
Another functional group of interest is the methoxy group present in isojamaicin (as shown in Fig. 2). The addition of a methoxy group (usually through chemical synthesis) has been done in several studies to determine if it contributes towards anti-cancer activity and to compare the difference in cytotoxic effects of the modified and unmodified drugs or compounds. In a study conducted by (Liew et al. 2020), methoxy groups were shown to contribute towards anti-metastatic activity using a derivative of quinolone and carboxamide. The simple addition of the functional group might not always favourable modify the compound. In a study conducted by Hille and Gust (2010) using [N,N′-bis(salicylidene)-1,2-phenylenediamine] iron(III) ([FeIII(salophene)Cl]), they examined the effects of the addition of a 3-methoxy and they indicated that the addition of the methoxy groups in the salicylidene rings resulted in the loss of tumour-cell selectivity. They also explained that shifting the position of the added methoxy groups contributed towards changes in cytotoxic effects. These studies indicate that the addition of methoxy groups may alter the pharmacological characteristics of pre-existing drugs or even unmodified compounds. The effects of this may indicate a difference in the cytotoxic activity regarding durmillone and isojamaicin but this will require further studies to investigate.
When examined with an AlamarBlue assay and in real-time using xCELLigence analysis, durmillone was shown to display concentration dependant variations in cytotoxicity which was shown in Figs. 3A, C and 4A, C which was also observed in a study conducted by Yan et al. (2019). Durmillone was also observed to be active against A549 cells in a concentration-dependent manner where higher concentrations of durmillone were inducing higher levels of apoptosis (Buyinza et al. 2021). There are no reported patterns of cytotoxicity of isojamaicin, however, in a study done by Buyinza et al. (2021), isojamaicin was shown to have cytotoxic activity against HEP G2 cells as well as DLD-1 WT and DLD-1 DKO colorectal cancer cells. As stated before, the study by Buyinza et al. (2021) showed that isojamiacin displayed cytotoxic activity against two DLD colorectal cell lines. They also showed that isojamaicin was inactive against A375 cells, indicating that the activity of isojamaicin is influenced by the cell type it is tested on. Isojamaicin displayed concentration-dependent cytotoxic activity on MIA PaCa 2 and HT 29 cells as shown in Figs. 3B, D and 4B, D.
Due to the pre-existing harmful side effects brought about by chemotherapy, anti-cancer drug development has to carefully consider the selective activity that potential anti-cancer drugs have towards cancer cells to protect normal body cells from damage and to protect a patient from experiencing harsh side effects (Liu et al. 2015). According to Buyinza et al. (2021), durmillone displayed extremely low and insignificant levels of cytotoxicity on BEAS-2B lung cells and no cytotoxicity on the CCD 19Lu lung fibroblast cells and LO2 liver cells. In the same study, isojamaicin showed levels of mild cytotoxicity on LO2 cells and no cytotoxicity on the BEAS-2B CCD 19Lu cells. This means that little or no cytotoxicity was expected to be seen on the HEK 293 cells following the treatment with isojamaicin and durmillone. Low and insignificant levels of cytotoxic activity were displayed in the HEK 293 cell line when treated with isojamaicin and durmillone as shown in Figs. 3E, F and 4E, F and the IC50s generated in both the AlamarBlue assay This means that compounds such as durmillone and isojamaicin which were selectively more toxic to MIA PaCa 2 and HT 29 are likely suitable candidates for further testing and possibly drug development.
Regarding the tests following AlamarBlue and xCELLigence, the IC50s generated from the xCELLignece analysis were used. The xCELLigence analysis can not only provide real-time quantitative analysis of cell activity but is also a label or stain-free system (Atmaca et al. 2016). This enables the calculation of time-dependent IC50s where an IC50 can be calculated based on where prominent activity is shown, making it a more accurate and informative IC50 (Atmaca et al. 2016). For this reason, the IC50s calculated by xCELLigence were used for further studies, rather than the AlamarBlue IC50s.
To restore cell machinery and halt cancer procession, apoptosis and cell cycle are important processes to be restored. Apoptosis is cell designed programmed cell death that should by its own character lead the cell into death should the cell be identified as infected or damaged. This process is controlled by many genes that acts as transcriptional factors for each other. Targeting defective apoptosis in cancerous cells has become a key element in cancer treatment; where it can be achieved by aiming to restore or bypass the defectives of the apoptosis pathway, which cancerous cells use to their advantage to survive (Wong 2011). Apoptosis detection was carried out for durmillone and isojamaicin treated MIA PaCa 2 and HT 29 cells using a caspase 3/7 assay and real-time PCR. Caspase activity was seen to be up-regulated in both MIA PaCa 2 and HT 29 after being treated with both durmillone and isojamaicin is seen to display a slightly higher level of caspase activity, compared to isojamaicin which suggests that the compounds tend to behave differently when used as a treatment on the same cell line. Caspase activity is characteristic of apoptosis, which suggests that these compounds induce apoptosis in cancer cells. Real-time PCR, which examined the gene expression levels of BAX, BCL-2 and p53 further verified the occurrence of apoptosis.
BCL-2 proteins are present in non-apoptotic cells to ensure that BAX proteins such as BCL2L4 do not accumulate and bind to the mitochondrial membrane leading to their permeabilisation (Karaliotas et al. 2015). The down-regulation of BCL-2 and up-regulation of BAX brought about by the treatment of durmillone and isojamaicin (Fig. 6) indicates that cytotoxicity of MIA PaCa 2 and HT 29 is brought about by the induction of apoptosis which can be verified further by the up-regulation of caspase 3 and 7 activity (Fig. 5).
The evident down-regulation of BCL-2 is important in determining whether or not compounds such as durmillone and isojamaicin hold the potential to be anti-cancer agents. One of the key strategies for anti-cancer drug development has been to control and terminate cancerous cells, by targeting the cell’s own mechanisms, in most cases, the components of the apoptotic pathway. The targeting of BCL-2 proteins and genes for inhibition can result in the induction of apoptosis in cancer cells. In the case of durmillone and isojamaicin, it is not known if the down-regulation of the BCL-2 gene is in conjunction with BCL-2 protein inhibition, and further studies are required to examine this concept.
p53 can be both an apoptosis-inducing gene or in cancer cells, it can be mutated and function as an oncogene. Both MIA PaCa 2 and HT 29 cells are cancers that contain mutated p53 genes (Deer et al. 2010, Candeias et al. 2016). p53 has been studied in cancer drug development through gene-based therapy and drug-based therapy. Gene-based therapy makes use of wild-type or unmutated p53 to destroy cells with deficient or defective p53 to trigger p53-mediated apoptosis or to sensitize cancerous cells to chemotherapeutic or radiation therapies (Wong 2011). Figure 6 has depicted the down-regulation of p53 in both the MIA PaCa 2 and HT 29 cell lines, following their treatment with durmillone and isojamaicin. This indicates that p53-mediated apoptosis does not occur and the compounds have no apparent influence on the restoration of mutated p53.
Conclusion
M. griffoniana contains a number of bioactive compounds and in this study, durmillone and isojamaicin were examined and found to display anti-cancer activity on MIA PaCa 2 and HT 29 cells. This was evident in both the AlamarBlue and xCELLigence cellular viability assays. The cytotoxic activity was brought about by apoptosis, which was evident from the up-regulation of caspase 3 and 7 activity, the up-regulation of the BAX gene as well as the down-regulation of BLC-2 and p53. Even though potential anti-cancer activity is displayed by these two compounds, further studies need to be done for further verification of their activity. This includes examining the functional differences of isojamaicin and durmillone regarding the addition of a methoxy group needs to be explored to determine how the methoxy group contributes towards isojamaicin’s bioactivity. Further studies also need to be done towards understanding the apoptotic mechanisms that these compounds act on to invoke their cytotoxic activity; including the study of other genes, proteins and receptors that these compounds may interact with in cancer cells. Mammalian studies should also be explored to obtain a better understanding of these compounds’ activity in a living organism, especially to obtain a better understanding if non-cancerous cells and organs can be affected by these compounds.
Data availability
All data is made available in the manuscript.
References
Atmaca H, Bozkurt E, Kısım A, Uslu R (2016) Comparative analysis of XTT assay and xCELLigence system by measuring cytotoxicity of resveratrol in human cancer cell lines. Turk J Biochem 41(6):413–421
Banzouzi JT, Prost A, Rajemiarimiraho M, Ongoka P (2008) Traditional uses of the African Millettia species (Fabaceae). Int J Bot 4:406–420
Buyinza D, Yang LJ, Derese S, Ndakala A, Coghi P, Heydenreich M, Wong VKW, Möller HM, Yenesew A (2021) Cytotoxicity of isoflavones from Millettia dura. Nat Prod Res 35(16):2744–2747
Candeias MM, Hagiwara M, Matsuda M (2016) Cancer-specific mutations in p53 induce the translation of Δ160p53 promoting tumorigenesis. EMBO Rep 17(11):1542–1551
Deer EL, González-Hernández J, Coursen JD, Shea JE, Ngatia J, Scaife CL, Firpo MA, Mulvihill SJ (2010) Phenotype and genotype of pancreatic cancer cell lines. Pancreas 39(4):425–435
DeVita VT, Lawrence TS, Rosenberg SA (2015) Cancer: principles & practice of oncology: primer of the molecular biology of cancer. Wolters Kluwer Health, Philadelphia, pp 1–7
Hanahan D, Weinberg R (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
Hille A, Gust R (2010) Influence of methoxy groups on the antiproliferative effects of [FeIII(salophene-OMe)cl] complexes. Eur J Med Chem 45(11):5486–5492
Karaliotas GI, Mavridis K, Scorilas A, Babis GC (2015) Quantitative analysis of the mRNA expression levels of BCL2 and BAX genes in human osteoarthritis and normal articular cartilage: An investigation into their differential expression. Mol med rep 12(3):4514–4521
Kumar D, Sharma P, Singh H, Nepali K, Gupta GK, Jain SK, Ntie-Kang F (2017) The value of pyrans as anticancer scaffolds in medicinal chemistry. RSC Adv 7(59):736977–36999
Liew SK, Malagobadan S, Arshad NM, Nagoor NH (2020) A review of the structure-activity relationship of natural and synthetic antimetastatic compounds. Biomolecules 10(1):138. https://doi.org/10.3390/biom10010138
Liu B, Ezeogu L, Zellmer L, Yu B, Xu N, Joshua Liao D (2015) Protecting the normal in order to better kill the cancer. Cancer Med 4(9):1394–1403
Mizrahi JD, Surana R, Valle JW, Shroff RT (2020) Pancreatic cancer. Lancet 395(10242):2008–2020
Ngamga D, Yankep E, Tane P, Bezabih M, Ngadjui BT, Fomum ZT, Abegaz BM (2005) Isoflavonoids from seeds of millettia griffoniana (bail) 15. Zeitschrift für Naturforschung B J Chem Sci 60(9):973–977
Rawla P, Sunkara T, Gaduputi V (2019) Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J Oncol 10(1):10–27
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2020) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249
Wong RSY (2011) Apoptosis in cancer: From pathogenesis to treatment. J Exp Clin Cancer Res CR 30(1):87
Xi Y, Xu P (2021) Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol 14(10):101174
Yan W, Yang J, Tang H, Xue L, Chen K, Wang L, Zhao M, Tang M, Peng A, Long C, Chen X, Ye H, Chen L (2019) Flavonoids from the stems of Millettia pachyloba drake mediate cytotoxic activity through apoptosis and autophagy in cancer cells. J Adv Res 20:117–127
Acknowledgements
The authors wish to acknowledge the university of Johannesburg and University of University of Yaoundé I for the use of their facilities and equipment to conduct the experiments behind this project. They also wish to acknowledge the staff members and students who provided assistance for the required lab work and experiments.
Funding
Open access funding provided by University of Johannesburg. Funding was received from the South African National Research Foundation NRF as a research grant awarded to Dr Mpho S. Choene.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval
This article does not contain any studies involving animals or humans performed by any of the authors.
Conflict of interest
Fatima Hoosen has no conflict of interest. Jordan Tonga Lembe has no conflict of interest. Derek Ndinteh has no conflict of interest. Tagatsing Fotsing Maurice has no conflict of interest. Yankep Emmanuel has no conflict of interest. Lesetja Raymond Motadi has no conflict of interest. Mpho Susan Choene has no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hoosen, F., Lembe, J.T., Ndinteh, D. et al. The anti-cancer potential of active compounds extracted from Millettia griffoniana on pancreatic and colorectal cancer cells. ADV TRADIT MED (ADTM) (2024). https://doi.org/10.1007/s13596-023-00733-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13596-023-00733-y