Abstract
Key message
Litter chemical quality regulates the distinct composition of the main microbial groups and ecoenzymatic stoichiometry. Microbes in spruce ( Picea asperata Mast.) and fir ( Abies faxoniana Rehd.) rather than birch ( Betula platyphylla Suk.) and rhododendron ( Rhododendron lapponicum (L.) Wahl.) can more easily adjust their physiological metabolism to acclimate to low N resources.
Context
Litter decomposition is the main pathway of nutrient cycling that bridges aboveground and underground material circulation and energy flow. Microorganisms are essential for the regulation of organic carbon decomposition and nutrient cycling.
Aims
We sought to reveal whether litter chemical quality predominates forest floor microbial structure and function in different species and how their characteristics vary with litter decomposition stages.
Methods
We measured litter substrate quality, microbial community structure, microbial biomass carbon (MBC) and nitrogen (MBN), extracellular enzyme activities and stoichiometric homeostasis of fresh litter (L), and fermentative (F) and humus (H) layers for these tree species.
Results
Overall, the enzyme activities and microbial biomass of birch and rhododendron were greater than those of spruce and fir. The microbial abundances of birch and rhododendron decreased with decomposition. Forest floor microbial nutrient limitation is generally restricted by N in subalpine forests, and ecoenzymatic stoichiometry is affected mainly by dissolved C/N/P stoichiometry. Stronger microbial C:N homeostasis (H′) was observed for spruce (5.56) and fir (4.17) than that for birch (1.82) and rhododendron (1.33).
Conclusion
We conclude that litter chemical quality led to the disparity in forest floor microbial groups and ecoenzymatic stoichiometry for different tree species.
Similar content being viewed by others
1 Introduction
Litter decomposition is the main pathway of nutrient return to soil and is a fundamental process in the forest ecosystem linking the aboveground and underground components of material circulation and energy flow (Berg and Mcclaugherty 2013; Maisto et al. 2011). Microorganisms are essential for the regulation of organic carbon decomposition, nitrogen mineralization, nutrient cycling, and transformation (Chapin et al. 2011; Attiwill and Adams 1993). The main microbial groups, bacteria, fungi, gram-negative bacteria (G−), gram-positive bacteria (G+), actinomycetes, protozoa, and some microalgae, are major decomposers in forest ecosystems by degrading complex organic compounds into simple small molecules (Ayres et al. 2006) and producing extracellular enzymes associated with carbon (C), nitrogen (N), and phosphorous (P) cycles (Waring 2013). Overall, microorganism biomass, activity, and community structure characteristics are closely related to the biochemical composition of litter (Cornwell et al. 2008; Genung et al. 2013; Chavez-Vergara et al. 2014).
The biochemical composition of litter is determined by the physiological characteristics of the plant species from which the material is derived. Litter of different tree species showed different characteristics during the decomposition processes (Ushio et al. 2008, 2010; Adamczyk et al. 2014). Litter substrate quality characteristics, such as C, N, ethanol-soluble compounds, dissolved organic matter, rates of C mineralization and net N mineralization, and denitrification potential, tend to differ between coniferous and hardwood species (Adamczyk et al. 2014). Such differences may be influenced by plant species and variations in the amount of plant biomass (Smolander et al. 2012). Furthermore, tree species can influence physico-chemical properties (Dan and Valentine 1991), microbial community composition (Ushio et al. 2008; Wang et al. 2015), and microbial biomass and enzyme activities (Grayston and Prescott 2005; Ushio et al. 2010; Zheng et al. 2017), and these effects have been attributed to species differences in litter quality, root exudates, and nutrient uptake (Hobbie 1992). Moreover, variation in tree species composition has been shown to affect structural and functional microbial community composition at both the within-stand (Saetre and Bååth 2000) and among-stand (e.g., Grayston and Prescott 2005) scales. The overall decomposition process can be viewed as the dynamics of fresh litter (L) being transformed into metastable humic substances through microbial and physical decay (Berg and Mcclaugherty 2013). In general, subalpine forest ecosystems have thick forest floors due to relatively slow decomposition (Fioretto et al. 2005). Litter decomposition has been divided into three stages based on analysis of the decomposition of Scots pine: early stage, late stage, and near-humus (H) stage (Berg and Mcclaugherty 2013). L, fermentative (F), and H layers have been simulated as three stages of litter decomposition to describe the litter decomposition process from L to the H stage, and litter substrate quality, main microbial groups, and ecoenzymatic stoichiometry change with the stage of decomposition during these processes (Ma et al. 2014).
Extracellular enzymes, biological catalysts produced by microorganisms, are thought to act as bottlenecks in ecologically important processes such as C, N, and P acquisition. Several ecoenzymes (β-glucosidase (BG), acid phosphatase (AP), leucine aminopeptidase (LAP), and β-N-acetylglucosaminidase (NAG)) have been identified as useful indicators of nutrient deficiency and microbial C, N, and P demand since they are major drivers of C and nutrient turnover in different ecosystems (Cui et al. 2018). Ecoenzymatic activity and microbial metabolism are integrated on the basis of resource demand relative to environmental availability (Sinsabaugh and Follstad Shah 2012). Ecoenzymatic stoichiometry connects the elemental stoichiometry of microbial biomass and detrital organic matter to microbial nutrient assimilation and growth. Therefore, ecoenzymes have been identified as useful indicators of nutrient deficiency and microbial nutrient demand. Microorganisms acclimate to stress by reassigning key resources to nutrient acquisition mechanisms rather than to growth (Cui et al. 2018). While it has also been reported that the ratio of C:N:P in microbial biomass is relatively conserved across ecosystems compared with the ratio in the litter, the microbial biomass ratio could indicate how allocation shifts alter nutrient demand (Cleveland and Liptzin 2007).
Stoichiometric homeostasis is the ability of organisms to maintain relatively stable chemical compositions regardless of environmental changes. Species with strong homeostasis play an important role in maintaining the microbial community structure, function, and stability of an ecosystem (Fanin et al. 2013; Li et al. 2016a, b). Microorganisms are the driving factors of the C, N, and P cycle at the ecosystem scale. The relationship between microbial biomass and nutrient stoichiometric characteristics determines the element cycling during litter decomposition (Spohn 2016). In turn, substrate quality and stoichiometry, especially available C, N, and P, affect microbial community structure and function (McDaniel et al. 2013). Our previous research found that litter quality drives the differentiation of microbial communities and ligninolytic and cellulolytic enzyme activities in the litter horizon across an alpine treeline ecotone (Zheng et al. 2018). In a subalpine fir forest, both microclimate and litter quality affect lignocellulose degradation and lignocellulolytic enzyme activities (Chen et al. 2018), and canopy gaps facilitate soil organic carbon (SOC) retention by soil microbial biomass in the organic horizon (Liu et al. 2018). However, litter decomposition in subalpine forest is a slow process and lacks long-term monitoring. Moreover, there is little evidence of major differences in the forest floor microbial structure and function at the tree species level, which has greatly restricted our understanding of the litter decomposition mechanism of subalpine forest ecosystems. Therefore, by means of space instead of time, we divide decomposition layers to simulate the different decomposition stages and study the forest floor microbial groups and ecoenzymatic stoichiometry between conifer and hardwood species.
In this study, we hypothesized that forest floor microbial community groups and ecoenzymatic stoichiometry vary in different tree species and regulated by litter chemical quality (C, N, P, dissolved organic carbon (DOC), dissolved nitrogen (DN), dissolved phosphorous (DP), and their stoichiometry). The main objective of this study was to examine the forest floor microbial community structure (using phospholipid fatty acid analysis—PLFA); microbial biomass carbon (MBC) and nitrogen (MBN) levels; extracellular enzyme activity and substrate quality in three layers (L, F, and H) under two coniferous species, spruce (Picea asperata Mast.) and fir (Abies faxoniana Rehd.); and two hardwood species, birch (Betula platyphylla Suk.) and rhododendron (Rhododendron lapponicum (L.) Wahl.), from subalpine forests on the eastern Tibetan Plateau. Specifically, we determine the following: (1) whether litter chemical quality of different tree species impacts forest floor main microbial groups and extracellular enzyme activity and (2) how characteristics of microbial structure and function vary with litter decomposition stages.
2 Materials and methods
2.1 Study site and sample collection
This study was conducted in the Miyaluo Nature Reserve (E102° 53′–102° 57′, N31° 14′–31° 19′, 2458–4619 m above sea level (a.s.l.)), Li County, Sichuan, Southwest China. The subalpine forest is located in the upper reaches of the Yangtze River and on the eastern Tibetan Plateau. This region contains broad-leaved forest, coniferous and broad-leaved mixed forest, dark coniferous forest, dwarf forest, and alpine shrub meadow from the valley to the hilltop. The alpine timberline occurs at approximately 4000 m, and the alpine desert occurs above 4500 m. The climate is cool in summer and cold in winter; the annual average temperature is 6–12 °C, the average temperature in January is − 8 °C, the average temperature in July is 12.6 °C, and the annual accumulated temperature is 1200–1400 °C. The annual precipitation varies from 600 to 1100 mm, and the annual evaporation is 1000–1900 mm from 2458 to 4619 m a.s.l. (Liu et al. 2016). Winter snow lasts from October to April of the following year, up to 6–7 months. The dominant tree species in the study area are Abies faxoniana Rehd., Betula platyphylla Suk., Betula albosinensis Burk., Picea asperata Mast., and Sabina saltuaria (Rehd. et Wils.). The dominant shrub species are Rhododendron lapponicum (L.) Wahl., Sorbus rufopilosa Schneid., Lonicera lanceolata Wall., and Salix paraplesia Schneid.et al. The dominant families of grasses are Gramineae, Compositae, and Gentianaceae (Liu et al. 2016).
Dominant subalpine forest species of spruce, Minjiang fir, white birch, and rhododendron in this region were selected (Table 4). The area of each forest stand was more than 1 ha, and the above dominant species constituted more than 90% of all tree species. The forest soil is an acid-udic cambisol (USDA soil taxonomic system), and the organic layer is a typical mor-type H. Samples were collected prior to the winter snow season in mid-October 2015. Three study stands (30 m × 30 m) per species were established, with a distance of at least 1000 m between stands. In each stand, five sample plots (1 m × 1 m) were randomly collected systematically from the forest floors from top to bottom at the same point, and at least 5 m between each plots. The forest layers were divided into L, F, and H per plot and layer. L consisted of fresh or slightly decomposed litter from trees and understory; F of partly decomposed litter, the origin of which was mainly identifiable; and H of decomposed organic matter, the origin of which could not be identified (Kanerva and Smolander 2007). After green plant material and large particles, such as cones and large roots, were removed, the L and F samples were cut into approximately 0.25-cm2 pieces; samples from H were sieved using a 2-mm sieve. Total number of samples were 180 (3 study stands × 4 forest species × 5 plots × 3 forest layers). Samples from each plot were divided into three subsamples: one was frozen at 4 °C until enzyme analysis within 1 week; a second was frozen at − 80 °C for PLFA analysis; and a third was dried at 65 °C to constant weight for chemical analyses.
2.2 Microbial biomass carbon and nitrogen
Microbial C and microbial N were estimated by chloroform fumigation–extraction (Ran and Liu 2009). The microbial biomass in the litter was determined according to differences between fumigated and unfumigated samples following extraction with 0.5 mol L−1 K2SO4. After shaking for 20 min, the extract was filtered, and MBC and MBN were determined by K2Cr2O7 oxidation-FeSO4 titration and the semi-micro Kjeldahl N method. The conversion factors for MBC and MBN were kMBC = 0.38 (Vance et al. 1987) and kMBN = 0.45 (Tan et al. 2014).
2.3 Phospholipid fatty acid analysis
We investigated PLFA composition both to evaluate changes in microbial biomass with the forest floor layer and tree species and as an index of the viability in the microbial community structure (Federle et al. 2011). PLFAs were extracted, fractionated, and methylated as described by Bossio and Scow (1998)). Individual fatty acid methyl esters (FAMEs) were identified by gas chromatography/mass spectrometry (GC/MS, Model QP-2010, Shimadzu, Japan). The fatty acid nomenclature used in this study was described by Frostegård et al. (1993a, b). The fatty acid 19:0 was added as an internal standard before methylation, and PLFAs were identified using the MIDI Sherlock Microbial Identification System (Version 132 4.5; MIDI, Newark, DE, 133 USA). Concentrations of individual fatty acids were determined as nanomoles per gram of dry matter (nmol g−1 DM). These tests used the PLFA tag (see Table 5). The ratios of fungi to bacteria (F:B) and of gram-positive bacteria to gram-negative bacteria (G+:G−) were calculated in this study.
2.4 Enzyme extraction and assays
Enzyme activity was determined after the extraction, determination, and preparation of crude enzyme solutions according to Allison et al. (Allison 2008). A total of 2 g of fresh sample was stirred for 2 min with 60 mL of acetic acid buffer solution (pH = 5) to obtain a crude enzyme solution. Assays were performed in ELISA plates in a 200-μL volume, with 50 μL of crude enzyme solution and 150 μL of buffer or 150 μL of substrate; for each sample, a control (crude enzyme+ buffer) and a test (crude enzyme+ substrate) were included. For PER, 10 μL of 0.3% H2O2 was added to the substrate control and sample groups. Samples were incubated as follows: AP for 45 min; peroxidase (POD) for 1–2 h; NAG for 3 h; cellobiohydrolase (CBH) for 4 h; BG for 1 h; and LAP for 4–6 h. After incubation, 5 μL of 1.0 M NaOH was added to all wells to terminate the reaction and develop color. Absorbance was measured at 405 nm for AP, NAG, CBH, LAP, and BG and at 450 nm for PPO and POD. The enzyme commission (EC) number, specific substrates, and substrate concentrations are provided in Table 6. Activity is expressed as micromole of substrate hydrolysed per hour per gram of dry organic matter (μmol h−1 gDOM−1).
2.5 Chemical analysis
The forest floor pH was measured using a milled litter-to-solution (water) ratio of 1:2.5 (w/v). The carbon (C) content of litter was determined by the dichromate oxidation-ferrous sulfate titration method according to Zhang et al. (1999). The nitrogen (N) content was determined using the semi-micro-Kjeldahl method, and the phosphorus (P) content was determined by Mo-Sb colorimetry (Ashagrie et al. 2005). The dissolved organic carbon (DOC) content was determined according to the method of Uselman et al. (2012), the dissolved nitrogen (DN) content by the Kjeldahl method, and the dissolved phosphorus (DP) content by Mo-Sb colorimetry (Uselman et al. 2012).
2.6 Stoichiometric homeostasis and threshold elemental ratio
-
(1)
EEAC:N = LnBG/Ln (LAP + NAG)
-
(2)
EEAC:P = LnBG/LnAP
-
(3)
EEAN:P = Ln (LAP + NAG)/LnAP
-
(4)
C:NR = DOC/DN
-
(5)
C:NB = MBC/MBN
-
(6)
H′ = 1/m
Equation (1) was used to calculate the ecoenzymatic carbon to nitrogen ratio, equation (2) was used to calculate the ecoenzymatic carbon to phosphorus ratio, and equation (3) was used to calculate the ecoenzymatic nitrogen to phosphorus ratio. The degree of microbial community-level C:N homeostasis (H′) was determined using Eq. (6) (Sterner and Elser 2002; Sinsabaugh et al. 2015). Stoichiometric homeostasis between the different decomposition stages for the tree species examined was assessed by regression analysis of the microbial biomass ratio and the soluble carbon and nitrogen ratio, where m is the slope of lnC:NR (resources) (4) versus lnC:NB (microbial biomass) (5), H′ > > 1 represents strong stoichiometric homeostasis, and H′ ≈ 1 represents weak or no homeostasis (Sterner and Elser 2002). There were no significant correlations between lnC:NR and lnC:NB (p > 0.05), which indicated strong community-level elemental homeostasis.
2.7 Statistical analysis
A mixed linear model ANOVA (SAS proc mixed) was applied to test the effects of forest layers and tree species on the substrate quality, enzyme activity, microbial biomass, and microbial communitieswith tree species as fixed main effects and forest layers as a random effect; post hoc multiple comparisons were employed to determine the variability in each indictor when testing statistically significant (p < 0.05). Pearson’s correlation analysis was used to evaluate relationships among enzyme activity, substrate quality, and stoichiometry. The microbial community structure was analysed by redundancy analysis (RDA) (length of gradient < 3 for microbial community variables) in R version 3.3.2 (using the vegan package; R Development Core Team, 2011), and the significance of the results was tested by a Monte Carlo permutation test (p < 0.001).
3 Results
3.1 Microbial community PLFA concentrations
The microbial community composition, including fungi, G+, G−, G−/G+, and total PLFA content, was significantly affected by both the tree species and forest layer (Fig. 1).
A mixed linear model for the effects of the forest layers and tree species on the microbial communities in litter. Post hoc multiple comparisons for the variability of microbial community determined as phospholipid acid (PLFA) concentrations in litter (L), fermentative (F), and humus (H) layers of the subalpine forests in China. Fresh litter (L), fermentative (F), and humus (H) layers. Different upper-case letters indicate significant differences among different forests within the same layer (p < 0.05). Different lower-case letters indicate significant differences between litter layers within the same forest type (p < 0.05). The same scheme is used in other figures
The microbial community composition differed significantly among tree species. A greater abundance of fungi biomass was observed in L under birch and rhododendron, whereas a higher G+ was found under spruce and fir. Compared with the other three species, G− and G−/G+ were significantly higher and G+ significantly lower in L for birch. In F, G−, G−/G+, and total PLFA contents for fir were higher than for the other species. The microbial community PLFA concentrations also differed among the forest floor layers. The number of fungi and G−, G−/G+, and total PLFA levels for birch and rhododendron decreased with decomposition. In addition, compared with the other layers, the contents of G+ and total PLFA for spruce were significantly greater in the H layer. In the forest floor under fir, bacteria, G−, G−/G+, and total PLFA all significantly increased and then decreased. In general, G+ decreased first and then increased for all tree species (Fig. 1).
3.2 Microbial biomass C and N
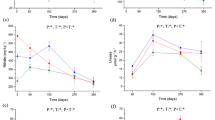
The forest layer and tree species significantly affected microbial C, microbial N, and their ratio (microbial C/microbial N) (Fig. 2). Overall, microbial C and microbial N levels for birch and rhododendron were higher than that of spruce and fir in F layer (Fig. 2). Microbial C continued to decrease with decomposition process, whereas microbial N first increased and then decreased. Overall, microbial C/microbial N decreased and was highest in the L layer.
A mixed linear model for the effects of the forest layers and tree species on the microbial biomass and their stoichiometry ratio. Post hoc multiple comparisons for the variability of microbial biomass carbon (MBC), microbial biomass nitrogen (MBN), and MBC/MBN in litter (L), fermentative (F), and humus (H) layers of the subalpine forests in China
3.3 Enzyme activity and ecoenzymatic stoichiometry
Enzyme activities and their stoichiometric ratios differed among tree species. The results of the mixed linear model showed that tree species and forest floors had significant effects on enzyme activities and ecoenzymatic stoichiometry (Fig. 3). Overall, AP, CBH, LAP, and NAG for birch and rhododendron were higher than those for spruce and fir in L and F and were particularly higher for birch. POD activity was higher for rhododendron than for the other species. EEAC:N was 0.86–1.30, EEAC:P was 0.63–1.07, and EEAN:P was 0.68–0.82. The ecoenzymatic stoichiometry of the forest floor was highest under spruce.
A mixed linear model for the effects of the forest layers and tree species on the enzyme activity. Post hoc multiple comparisons for the variability of enzyme activity (β-glucosidase (BG), acid phosphatase (AP), cellobiohydrolase (CBH), leucine aminopeptidase (LAP), β-N-acetylglucosaminidase (NAG), and POD (peroxidase) in litter (L), fermentative (F), and humus (H) layers of the subalpine forests in China
Enzyme activities also differed according to the decomposition stage. With ongoing decomposition, AP, BG, CBH, LAP, and NAG decreased for birch, fir, and spruce, whereas they initially increased and then decreased for rhododendron. POD activity increased under fir, spruce, and rhododendron but decreased under birch (Fig. 3). Overall, the EEAC:P and EEAN:P of all tree species decreased with the decomposition process (Fig. 4).
A mixed linear model for the effects of the forest layers and tree species on the ecoenzymatic stoichiometry. Post hoc multiple comparisons for the variability of ecoenzymatic stoichiometry ratio in litter (L), fermentative (F), and humus (H) layers of the subalpine forests in China. EEAC:N: LnBG/Ln (LAP + NAG), EEAC:P:LnBG/LnAP, EEAN:P: Ln (LAP + NAG)/LnAP
3.4 Substrate quality and C/N/P stoichiometry
Tree species and forest layer had significant effects on pH, C, N, P, DOC, DN, and DP as well as dissolved C/N/P stoichiometric (Table 1).
Compared with tree species, the pH values in the forest floors under spruce and fir were significantly higher than those under birch and rhododendron (Table 1). Additionally, the C, N, DOC, DN, and DP contents were highest for birch, especially in the L layer, and these contents were generally higher for birch and rhododendron than for spruce and fir. C/N was consistently higher under rhododendron than under birch, fir, and spruce. DN/DP was lowest in the forest floor under spruce (Fig. 5).
A mixed linear model for the effects of the forest layers and tree species on the stoichiometric ratio. Post hoc multiple comparisons for the variability of the carbon (C), nitrogen (N), and phosphorous (P) stoichiometric ratio and dissolved organic carbon (DOC), dissolved nitrogen (DN), and dissolved phosphorous (DP) stoichiometric ratio in litter (L), fermentative (F), and humus (H) layers of the subalpine forests in China
C, N, P, DOC, DN, and DP contents differed in three forest floor layers. N and DN contents for spruce and fir continued to decrease with ongoing decomposition, whereas those for birch and rhododendron first increased and then decreased. L contained more C, DOC, and DP than did the other layers. Overall, C/N and C/P showed a decreasing trend; DN/DP exhibited an increasing trend as decomposition continued.
3.5 Pearson correlation of forest floor microbial biomass, ecoenzymatic stoichiometry and litter chemical quality
Pearson correlation was used to assess the correlation between microbial biomass, enzyme activity, and stoichiometry with substrate quality and C/N/P stoichiometry. Based on the Pearson correlation, the microbial C, microbial N, and enzyme activities, except for that of POD, were positively correlated with C, N, DOC, DN, and DP. Conversely, POD was negatively correlated with C, DOC, DP, and BG (Table 2).
MBC/MBN was positively correlated with C/N and DOC/DN but negatively correlated with N/P and DN/DP. Moreover, EEAC:P was positively correlated with C/P and DOC/DN but negatively correlated with DOC/DP and DN/DP, and EEAC:N was positively correlated with DOC/DN but negatively correlated with DN/DP; EEAN:P was negatively correlated with DOC/DP and DN/DP (Table 3).
3.6 Redundancy analysis (RDA) of microbial community structure, enzyme activity, and substrate quality
RDA was used to assess the correlation between microbial community structure and enzyme activity and substrate quality. Based on RDA, the first two axes captured 41.4% of the variability in the microbial community structure, whereas RDA1 (the x-axis) and RDA2 (the y-axis) accounted for 28.1% and 13.3% of the variation, respectively. Among them, the major microbial variables were fungi, G−, G+, G−/G+, microbial biomass, substrate quality (except P), and enzyme activities (except POD), which contributed most to the separation of the samples. The amount of fungi and bacteria, F:B, and total PLFAs correlated positively with DOC, DP, BG, C, microbial C, microbial N, LAP, CBH, and DN but negatively with POD. G− and G−/G+ correlated positively with enzyme activities (except POD), microbial biomass, and substrate quality, and negatively with POD. G+ correlated negatively with microbial C, LAP, CBH, DN, NAG, AP, and N.
3.7 Stoichiometric homeostasis
To evaluate the strength of stoichiometric H′, we analysed associations between microbial biomass elemental ratios and C/N/P ratios in litter resources. All H′ values were greater than 1 (Fig. 7). LnC:NB for birch was linearly related to LnC:NR. Furthermore, the H′ values for spruce and fir were 5.56 and 4.17 and far greater than 1, whereas those of rhododendron and birch were 1.82 and 1.33 and close to 1 (Fig. 7). Overall, the linear regression relationship between LnC:NB and LnC:NR for fir, spruce and rhododendron was not significant (p > 0.05), although a significant linear correlation was observed for birch (p < 0.01).
4 Discussion
4.1 Differences among tree species
Awareness of the litter decomposition process of subalpine forest ecosystems requires an understanding of microbial community activity and ecoenzymatic stoichiometry at the tree species level. The results of our study suggest that litter chemical quality regulates the distinct composition of the main microbial groups and ecoenzymatic stoichiometry. Microorganisms represent essential components of the biotic system in natural forests because they are key players in nutrient turnover (Hackl et al. 2004). Some research has also found that the activity of the soil microbial community can be strongly affected by the biochemical composition of litter (Chavez-Vergara et al. 2014), which is consistent with the findings of Grayston (Grayston and Prescott 2005) and Adamczyk et al. (2014). In the present study, there was a greater biomass of fungi under birch and rhododendron at the early decomposition stages but with fewer fungi in late decomposition stages compared with spruce and fir. According to Li et al. (2017), fungi can be divided into functional groups that decompose different types of organic matter: fast-growing r-strategists specialized in the utilization of easily available resources and slow-growing k-strategists able to decompose more recalcitrant compounds. Fast-growing opportunistic fungi are stimulated by easily accessible C sources and high N availability (Chigineva et al. 2009), which may also be the reason why more fungi were found under birch and rhododendron than under spruce and fir in early decomposition stages but fewer were found at later stages. However, there were no significant differences in the forest floor fungi: bacteria (F:B) biomass between tree species, which indicates that the microbial community structure was relatively stable. F:B can often be used as an important indicator of overall structural changes in microbial communities (Frostegård et al. 1993a, b).
In general, forest floor enzyme activities and microbial biomass of birch and rhododendron were greater than those of spruce and fir (Figs. 2 and 3). This is in accordance with many studies (Priha and Smolander 1997; Priha et al. 2001; Smolander and Kitunen 2002; Kanerva and Smolander 2007). To a certain extent, microbial biomass not only represents the number and activity of microorganisms in the litter decomposition process but also acts as an important component of active organic carbon and nutrient pools (Lavelle 2000). The final decomposition of litter is completed under the combination of microorganisms and soil enzyme systems (Cleveland and Liptzin 2007). Climatic and edaphic variables directly and indirectly correlate with forest type and greatly impact enzyme activities (Zheng et al. 2017). Overall, the contents of lignin/N, terpenoids, and phenolic substances in conifer forests are relatively higher than those in hardwood forests, a situation that is not conducive to the growth of microbial communities and leads to relatively slow decomposition (Dai et al. 2001). The initial C/N ratio is an important indicator of litter decomposition that plays an important role in predicting the litter decomposition rate (Taylor et al. 2002). The rhododendron C/N was generally higher than that of birch, spruce, and fir (Fig. 5), constricting the decomposition rate and leading to the accumulation of recalcitrant materials.
In our study, the pH of the forest floor under spruce and fir was higher than that under birch and rhododendron (Table. 1), which is consistent with the research of Uselman et al. (2012). Sinsabaugh et al. (2008) suggested that pH has direct effects on the activity of extracellular enzymes, as most enzymes have an optimal pH, and Xiao et al. (2016) found DOC and DN to be negatively correlated, indicating that a lower pH will significantly increase the content of soluble organic matter. Such an increase in acidity will accelerate the leaching of multivalent cations, such as Al3+, Ca2+, which is beneficial for enhancing the solubility of organic matter, resulting in the organic matter content of birch and rhododendron being greater than that of spruce and fir. Therefore, birch and rhododendron may provide more nutrients for microbial metabolism and lead to differences in enzyme activity.
A correlation matrix and RDA suggested that the relationships between litter enzyme activity, microbial biomass, and microbial community can be best interpreted by C and N, but not P, as well as DOC, DN, and DP (Table 2), which is consistent with the findings of Li et al. 2016a, b). In our study, tree species significantly affected the litter chemical quality in the decomposition process (Table 1), and the C, N, DOC, DN, and DP contents for birch and rhododendron were higher than those for spruce and fir. Under established climate conditions, the microbial community is first regulated by the quantity and quality of available resources and then by physiological and chemical factors (such as pH, EC, salinity, and heavy metals), temperature, and humidity (Aanderud et al. 2008). Indeed, tree species inevitably affect the content and spatial distribution of enzyme activity and the microbial community by altering the chemical composition of leaf litter (Kanerva and Smolander 2007). Cui et al. (2018) found that variations in soil enzyme activities were best accounted for by vegetation characteristics and physicochemical properties. It is generally believed that litter enzyme activity is closely related to the contents of C, N, P, lignin, and cellulose, as well as to C/N, C/P, and lignin/N, among others (Criquet et al. 2004; Sinsabaugh et al. 2002). Therefore, enzyme activities and microbial biomasses change with litter nutrient contents. Accordingly, higher enzyme activities and microbial biomasses are associated with greater C, N, DOC, DN, and DP contents. Overall, hardwood litter contains more nutrients that are conducive to microbial activities; conversely, conifer litter has a low water content and more AIR such as lignin, cellulose, and tannin that are not conducive to the growth of microorganisms (Zhang et al. 2007). Greater available C/N/P of litter favors the growth of microbial populations and the accumulation of microbial biomass (Barbhuiya et al. 2004).
4.2 Differences among forest floor layers
Litter chemical quality and C/N/P stoichiometry regulate the distinct composition of the main microbial groups, as they represent different stages of decomposition and thus provide different substrates for decomposer organisms. Our results showed that microbial community structure and enzyme activities vary with forest floor layers. Overall, microbial C, AP, BG, CBH, LAP, and NAG continued to decrease with decomposition under spruce, fir, and birch, although microbial biomass and enzyme activity peaked in the F stage under rhododendron. Šnajdr et al. (2008) also found that microbial biomass and enzyme activities decreased with the forest soil profile and that the activities of all tested enzymes exhibited high spatial variability in the L and H horizons. Microbial groups display preferences for different carbon sources at different stages of litter decomposition, with bacteria readily decomposing newly entering carbon sources and fungi preferring hard-to-decompose, recalcitrant carbon sources (Andresen et al. 2014). G+ was significantly negatively affected by MBN (Fig. 6), which may be the reason why G+ first decreased and then increased.
Redundancy analysis (RDA) of microbial indices (blue lines) and environmental variables (red lines) with the ordination scores of seven biomarker classes. Seven biomarker classes were used to construct statistical matrix. The angle and length of the blue (and/or red) arrows (vectors) indicate the direction and strength of the relationship of microbial indices with the ordination scores in litter substrate quality (carbon (C), nitrogen (N) and phosphorous (P) stoichiometric ratio and dissolved organic carbon (DOC), dissolved nitrogen (DN), dissolved phosphorous (DP)) and enzyme activities (β-glucosidase (BG), acid phosphatase (AP), cellobiohydrolase (CBH), leucine aminopeptidase (LAP), β-N-acetylglucosaminidase (NAG) and POD (peroxidase)). Gram-positive bacteria (G+), gram-negative bacteria (G−), fungi: bacteria ratio (F/B), gram-negative:gram-positive (G−/G+) (blue lines). Trilateral: spruce; square: fir; rhombus: birch; circle: rhododendron. Yellow: fresh litter layer; white: fermentative layer; green: humus layer
Overall, the C, DOC, DP, and C/N contents for the four species and the N and DN contents for the spruce and fir decreased with decomposition, leading to a general trend of decline in microorganism abundance because the availability of microbial substrates was poor (Vance et al. 1987). This may be the reason for fungal degradation via decomposition.
Rapid leaching of L produced a large number of water-soluble substances and organic-soluble components in the early stage of decomposition, providing energy and nutrients for the growth and reproduction of microorganisms. Therefore, the biomass and activity of microorganisms were highest in the L layer. The main remaining fractions in the late stage of decomposition were insoluble, and microbial biomass and activity decreased significantly with litter layer during decomposition (Aerts 1997). Overall, the C/N of the four tree species consistently exhibited a gradual decrease with ongoing decomposition; mainly during the early stage of decomposition, the microbial matter involved a high degree of carbon source and organic matter utilization, which may lead to a rapid release of carbon (Aerts 1997), thereby reducing C/N and C/P. This might be the reason why microbial C, which mainly expresses the amount of living microorganisms present during litter decomposition (Larsen et al. 2002), decreased, and RDA indicated that the variation in microbial community structure during decomposition was mainly affected by microbial C (Wang et al. 2015). Microbial biomass pools and their stoichiometry are significantly and positively correlated with SOC (Liu et al. 2018), and decomposition progression is positively correlated with enzyme activities. POD degrades H and lignin (Simoes et al. 1997), and Preston et al. (2009) found that increased resistance to litter decomposition was due to the accumulation of insoluble and irregular high-molecular-weight structures (including condensed tannins, keratin, lignin, or modified lignin). Therefore, POD activity under spruce, fir, and rhododendron increased with decomposition, especially in the H layer of rhododendron. This may be because rhododendron contributes high amounts of lignin to the late stage of decomposition (Deng et al. 2015).
4.3 Ecoenzymatic stoichiometry and microbial stoichiometric homeostasis
The elemental stoichiometry of microorganisms is fundamental for understanding the production dynamics and biogeochemical cycles of ecosystems (Sinsabaugh et al. 2009). Macroecological studies show that the most widely measured ecoenzymatic activities have a similar stoichiometry for all microbial communities (Sinsabaugh et al. 2009; Sinsabaugh and Follstad Shah 2012). C-acquisition enzymes have scaling relationships similar to those of N- and P-acquisition enzymes (Tapia-Torres et al. 2015). At the global scale, EEAC:N:P showed 1:1:1, and EEAC:N:P was similar across ecosystems (Sinsabaugh et al. 2009). In the present study, EEAC:N was 0.86–1.30 (less than the global average), EEAC:P was 0.63–1.07 (more than the global average), and EEAN:P was 0.68–0.82 (also more than the global average). It was found that the ecoenzymatic stoichiometry of alpine forest showed relative specificity. Low EEAC:P and EEAN:P ratios indicate that the ecosystem nutrient cycle is restricted by P, and low EEAC:N and high EEAN:P ratios indicate that the ecosystem nutrient cycle is restricted by N (Waring et al. 2014). This means that the forest floor had relatively high activity of extracellular enzymes related to N degradation and low activity of P extracellular enzymes, which indicated that litter decomposition was limited by N in this subalpine forest. The findings may also reflect the relative lack of N in the alpine region, requiring microbes to produce more enzymes related to N release to meet their metabolic needs.
The results showed that ecoenzymatic stoichiometry was affected mainly by dissolved C/N/P stoichiometry rather than by C/N/P stoichiometry. Moreover, the specific performance of ecoenzymatic stoichiometry correlated positively with DOC/DN and negatively with DN/DP (Table. 3). McDaniel et al. (2013) found that substrate quality and stoichiometry, especially soluble C/N/P, affect microbial community structure and function. Additionally, enzyme activity and chemical stoichiometry are controlled by C, N, and P resources (Sinsabaugh et al. 2009; Sinsabaugh et al. 2015). As mentioned above, microbial activity and function also largely depend on the water content, and the absorbance of enzymes by litter may result in variation in ecoenzymatic ratios (Romanowicz et al. 2016). Therefore, the results of this study suggest that the chemical quality of litter is a factor strongly impacting ecoenzymatic stoichiometry and nutrient availability, which mostly affects microbial nutrient ratios. For instance, DOC/DN had a strong influence on MBC/MBN in the decomposition process under birch, with up to 47% of the variation. Moreover, forest floor microbial stoichiometric homeostasis was far greater than 1 for spruce and fir, whereas values for birch and rhododendron were close to 1 (Fig. 7). These findings suggest that the chemical balance of microbes in the decomposition process varies greatly between different species and that the microbial communities under conifers have stronger homeostasis. Accordingly, microbes under spruce and fir can adjust their physiological metabolism to require low levels of N and P resources, thereby acclimating to oligotrophic habitats (Cui et al. 2018).
5 Conclusions
In summary, litter chemical quality led to the disparity in forest floor microbial groups and ecoenzymatic stoichiometry between different trees pecies. The microbial community PLFA biomass, microbial C, microbial N, and ecoenzyme activities changed with litter decomposition stages. Compared with ecoenzymatic stoichiometry at the global scale, microbial nutrient limitation is restricted by N in subalpine forests, and ecoenzymatic stoichiometry is mainly affected by dissolved C/N/P stoichiometry rather than by C/N/P stoichiometry. Furthermore, we speculate that microbes under spruce and fir can more easily adjust and acclimate to their environment in terms of global change, because microbial communities under spruce and fir have stronger H′ than do those under birch and rhododendron. These observations will provide a theoretical basis for understanding the regional nutrient cycling process linking plants and microbes in subalpine forest ecosystems.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Aanderud ZT, Shuldman MI, Drenovsky RE et al (2008) Shrub-interspace dynamics alter relationships between microbial community composition and belowground ecosystem characteristics [J]. Soil Biol Biochem 40(9):2206–2216
Adamczyk B, Kilpeläinen P, Kitunen V et al (2014) Potential activities of enzymes involved in N, C, P and S cycling in boreal forest soil under different tree species [J]. Pedobiologia 57(2):97–102
Aerts R (1997) Climate, leaf litter chemistry and leaf litter decomposition in terrestrial ecosystems: a triangular relationship [J]. Oikos 79(79):439–449
Allison S (2008). Colorimetric enzyme assays. Allison Lab Protocol.
Andresen LC, Dungait JA, Bol R et al (2014) Bacteria and fungi respond differently to multifactorial climate change in a temperate heathland, traced with 13C-glycine and FACE CO2 [J]. PLoS One 9(1):e85070
Ashagrie Y, Zech W, Guggenberger G (2005) Transformation of a Podocarpus falcatus, dominated natural forest into a monoculture Eucalyptus globulus, plantation at Munesa, Ethiopia: soil organic C, N and S dynamics in primary particle and aggregate-size fractions [J]. Agric Ecosyst Environ 106(1):89–98
Attiwill PM, Adams MA (1993) Nutrient cycling in forests [J]. New Phytol 124(4):561–582
Ayres E, Dromph KM, Bardgett RD (2006) Do plant species encourage soil biota that specialise in the rapid decomposition of their litter? [J]. Soil Biol Biochem 38(1):183–186
Barbhuiya AR, Arunachalam A, Pandey HN et al (2004) Dynamics of soil microbial biomass C, N and P in disturbed and undisturbed stands of a tropical wet-evergreen forest [J]. Eur J Soil Biol 40(3–4):113–121
Berg B, Mcclaugherty C (2013). Plant Litter. Decomposition, humus formation, carbon sequestration [M]. Berlin:Springer.
Bossio DA, Scow KM (1998) Impacts of carbon and flooding on soil microbial communities: phospholipid fatty acid profiles and substrate utilization patterns [J]. Microb Ecol 35(3-4):265–278
Cao Y, Li Y, Li C et al (2016) Relationship between presence of the desert shrub Haloxylon ammodendron, and microbial communities in two soils with contrasting textures [J]. Appl Soil Ecol 103:93–100
Chapin FS, Matson PA, Mooney HA (2011) Principles of terrestrial ecosystem ecology. Springer, Berlin
Chavez-Vergara B, Merino A, Vázquez-Marrufo G et al (2014) Organic matter dynamics and microbial activity during decomposition of forest floor under two native neotropical oak species in a temperate deciduous forest in Mexico[J]. Geoderma 2014(235-236):133–145
Chen YM, Liu Y, Zhang J et al (2018) Microclimate exerts greater control over litter decomposition and enzyme activity than litter quality in an alpine forest-tundra ecotone [J]. Sci Rep 8(1)
Chigineva N, Aleksandrova A, Tiunov A (2009) The addition of labile carbon alters litter fungal communities and decreases litter decomposition rates [J]. Appl Soil Ecol 42:264–270
Cleveland CC, Liptzin D (2007) C:N:P stoichiometry in soil: is there a “Redfield ratio” for the microbial biomass? [J]. Biogeochemistry 85(3):235–252
Cornwell WK, Cornelissen JHC, Amatangelo K et al (2008) Plant species traits are the predominant control on litter decomposition rates within biomes worldwide [J]. Ecol Lett 11(10):1065–1071
Criquet S, Ferre E, Farnet AM et al (2004) Annual dynamics of phosphatase activities in an evergreen oak litter: influence of biotic and abiotic factors [J]. Soil Biol Biochem 36(7):1111–1118
Cui Y, Fang L, Guo X et al (2018) Ecoenzymatic stoichiometry and microbial nutrient limitation in rhizosphere soil in the arid area of the northern Loess Plateau, China [J]. Soil Biol Biochem 116:11–21
Dai L, Xu Z, Zhang Y et al (2001) Study on decomposition rate and fall of Pinus koraiensis needle [J]. Acta Ecol Sin 21(8):1296–1300
Dan B, Valentine D (1991) Fifty-year biogeochemical effects of green ash, white pine, and Norway spruce in a replicated experiment [J]. Forest Ecol Manag 40(1-2):13–25
Deng CC, Jiang XM, Liu Y et al (2015) Litter decomposition of Rhododendron lapponicum in alpine timberline ecotone [J]. Acta Ecol Sin 35(6):1769–1778
Fanin N, Fromin N, Buatois B, Hättenschwiler S (2013) An experimental test of the hypothesis of non-homeostatic consumer stoichiometry in a plant litter-microbe system[J]. Ecol Lett 16(6):764–772
Federle TW, Livingston RJ, Wolfe LE et al (2011) A quantitative comparison of microbial structure of estuarine sediments form microcosms and the field [J]. Can J Microbiol 32(4):319–325
Fioretto A, Nardo CD, Papa S et al (2005) Lignin and cellulose degradation and nitrogen dynamics during decomposition of three leaf litter species in a Mediterranean ecosystem [J]. Soil Biol Biochem 37(6):1083–1091
Frostegård A, Tunlid A, Bååth E (1993a) Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals [J]. Appl Environ Microbiol 59(11):3605–3617
Frostegård A, Bååth E, Tunlio A (1993b) Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis [J]. Soil Biol Biochem 25(6):723–730
Genung MA, Bailey JK, Schweitzer JA (2013) The afterlife of interspecific indirect genetic effects: genotype interactions alter litter quality with consequences for decomposition and nutrient dynamics [J]. PLoS One 8
Grayston SJ, Prescott CE (2005) Microbial communities in forest floors under four tree species in coastal British Columbia [J]. Soil Biol Biochem 37(6):1157–1167
Hackl E, Bachmann G, Zechmeisterboltenstern S (2004) Microbial nitrogen turnover in soils under different types of natural forest [J]. Forest Ecology & Management 188(1–3):101–112
Hobbie SE (1992) Effects of plant species on nutrient cycling [J]. Trends Ecol Evol 7(10):336–339
Kanerva S, Smolander A (2007) Microbial activities in forest floor layers under silver birch, Norway spruce and Scots pine [J]. Soil Biol Biochem 39(7):1459–1467
Larsen KS, Jonasson S, Michelsen A (2002) Repeated freeze-thaw cycles and their effects on biological processes in two arctic ecosystem types [J]. Appl Soil Ecol 21(3):187–195
Lavelle P (2000) Ecological challenges for Soil science[J]. Soil Sci 165(1):73–86
Li S, Wang Z, Yang J (2016a) Changes in soil microbial communities during litter decomposition [J]. Biodivers Sci 24(2):195–204
Li Y, Li Q, Guo D et al (2016b) Ecological stoichiometry homeostasis of Leymus chinensis in degraded grassland in western Jilin Province, NE China[J]. Ecol Eng 90:387–391
Li Y, Li Y, Chang SX et al (2017) Linking soil fungal community structure and function to soil organic carbon chemical composition in intensively managed subtropical bamboo forests [J]. Soil Biol Biochem 107:19–31
Liu Y, Chen Y, Zhang J et al (2016) Changes in foliar litter decomposition of woody plants with elevation across an alpine forest–tundra ecotone in eastern Tibet Plateau [J]. Plant Ecol 217(5):495–504
Liu Y, Zhang J, Yang W et al (2018) Canopy gaps accelerate soil organic carbon retention by soil microbial biomass in the organic horizon in a subalpine fir forest [J]. Appl Soil Ecol 125:169–176
Ma WJ, Zhao YT, Zhang QQ et al (2014) C: N: P stoichiometry in forest floor litter of evergreen broad-leaved forests at different successional stages in Tiantong, Zhejiang, eastern China [J]. Chin J Plant Ecol 38(8):833–842
Maisto G, De MA, Meola A et al (2011) Nutrient dynamics in litter mixtures of four Mediterranean maquis species decomposing in situ [J]. Soil Biol Biochem 43(3):520–530
McDaniel, M.D., Kaye, J.P. and Kaye, M.W. 2013 Increased temperature and precipitation had limited effects on soil extracellular enzyme activities in a post-harvest forest. Soil Biol Biochem 56:90-98
Preston CM, Nault JR, Trofymow JA et al (2009) Chemical changes during 6 years of decomposition of 11 litters in some canadian forest sites. Part 1. Elemental Composition, Tannins, Phenolics, and Proximate Fractions [J]. Ecosystems 12(7):1053–1077
Priha O, Smolander A (1997) Microbial biomass and activity in soil and litter under Pinus sylvestris, Picea abies and Betula pendula at originally similar field afforestation sites [J]. Biol Fertil Soils 24(1):45–51
Priha O, Grayston SJ, Hiukka R et al (2001) Microbial community structure and characteristics of the organic matter in soils under Pinus sylvestris, Picea abies and Betula pendula at two forest sites.[J]. Biol Fertil Soils 33(1):17–24
Ran H, Liu QB (2009) Effects of seasonal freezing-thawing on microbial count and biomass in Abies faxoniana and Betula platyphylla Litters [J]. J Sichuan Agric Univ 27(4):450–454
Romanowicz KJ, Freedman ZB, Upchurch RA et al (2016) Active microorganisms in forest soils differ from the total community yet are shaped by the same environmental factors: the influence of pH and soil moisture [J]. FEMS Microbiol Ecol 92(10)
Saetre P, Bååth E (2000) Spatial variation and patterns of soil microbial community structure in a mixed spruce-birch stand.[J]. Soil Biol Biochem 32(7):909–917
Simoes DDCM, Mcneill D, Kristiansen B et al (1997) Purification and partial characterisation of a 1.57 kDa thermostable esterase from Bacillus stearothermophilus [J]. FEMS Microbiol Lett 147(1):151–156
Sinsabaugh RL, Follstad Shah JJ (2012) Ecoenzymatic stoichiometry and ecological theory[J]. Annu Rev Ecol Evol Syst 43(1):313–343
Sinsabaugh RL, Carreiro MM, Repert DA (2002) Allocation of extracellular enzymatic activity in relation to litter composition, n deposition, and mass loss [J]. Biogeochemistry 60(1):1–24
Sinsabaugh RL, Lauber CL, Weintraub MN et al (2008) Stoichiometry of soil enzyme activity at global scale.[J]. Ecol Lett 11(11):1252
Sinsabaugh RL, Hill BH, Shah JJF (2009) Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment [J]. Nature 462(7274):795–798
Sinsabaugh RL, Shah JJF, Findlay SG et al (2015) Scaling microbial biomass, metabolism and resource supply [J]. Biogeochemistry 122(2-3):175–190
Smolander A, Kitunen V (2002) Soil microbial activities and characteristics of dissolved organic C and N in relation to tree species [J]. Soil Biol Biochem 34(5):651–660
Smolander A, Kanerva S, Adamczyk B et al (2012) Nitrogen transformations in boreal forest soils—does composition of plant secondary compounds give any explanations?[J]. Plant Soil 350(1-2):1–26
Šnajdr J, Valášková V, Merhautová V et al (2008) Spatial variability of enzyme activities and microbial biomass in the upper layers of Quercus petraea forest soil [J]. Soil Biol Biochem 40(9):2068–2075
Spohn M (2016) Element cycling as driven by stoichiometric homeostasis of soil microorganisms [J]. Basic Appl Ecol S1439179116300391
Sterner RW, Elser JJ (2002). Ecological stoichiometry: the biology of elements from molecules to the biosphere [M]// Ecological Stoichiometry: the Biology of Elements from Molecules to the Biosphere 225-226.
Tan B, Wu FZ, Qin JL, Wu QG, Yang WQ (2014) Dynamics of soil microbial biomass and enzyme activity in the subalpine/alpine forests of western Sichuan [J]. Ecol Environ Sc 23(8):1265–1271
Tapia-Torres Y, Elser JJ, Souza V et al (2015) Ecoenzymatic stoichiometry at the extremes: how microbes cope in an ultra-oligotrophic desert soil [J]. Soil Biol Biochem 87:34–42
Taylor JP, Wilson B, Mills MS, Burns RG (2002) Comparison of microbial numbers and enzymatic activities in surfacesoils and subsoils using various techniques [J]. Soil Biol Biochem 34(3):387–401
Team R D C. R Reference Manual - Volume 4 - Methods and Tools - for R version 2.13[M]. 2011
Uselman SM, Qualls RG, Lilienfein J (2012) Quality of soluble organic C, N, and P produced by different types and species of litter: root litter versus leaf litter [J]. Soil Biol Biochem 54(6):57–67
Ushio M, Wagai R, Balser TC et al (2008) Variations in the soil microbial community composition of a tropical montane forest ecosystem: does tree species matter? [J]. Soil Biol Biochem 40(10):2699–2702
Ushio M, Kitayama K, Balser TC (2010) Tree species effects on soil enzyme activities through effects on soil physicochemical and microbial properties in a tropical montane forest on Mt. Kinabalu, Borneo [J]. Pedobiologia 53(4):227–233
Vance ED, Brookes PC, Jenkinson DS (1987) An extraction method for measuring soil microbial biomass C [J]. Soil Biol Biochem 19(6):703–707
Wang H, Liu S, Chang SX et al (2015) Soil microbial community composition rather than litter quality is linked with soil organic carbon chemical composition in plantations in subtropical China [J]. J Soils Sediments 15(5):1094–1103
Waring BG (2013) Exploring relationships between enzyme activities and leaf litter decomposition in a wet tropical forest[J]. Soil Biol Biochem 64(9):89–95
Waring BG, Weintraub SR, Sinsabaugh RL (2014) Ecoenzymatic stoichiometry of microbial nutrient acquisition in tropical soils. Biogeochemistry 117(1):101–111
Xiao HY, Liu B, Yu ZP, Wan XH, Sang CP, Zhou FW, Huang ZQ (2016) Effects of forest types on soil dissolved organic carbon and nitrogen in surface and deep layers in subtropical region, China.[J]. Chin J Appl Ecol 27(4):1031–1038
Zhang WR, Yang GY, Tu XN, et al (1999). Determination of organic matter in forest soil and calculation carbon-nitrogen ratio[S]. Forestry industry standard of the People's Republic of China 106-108.
Zhang P, Tian XJ, He XB et al (2007) Enzyme activities in litter, fragmentation and humus layers of subtropical forests[J]. Ecol Environ 16(3):1024–1029
Zheng HF, Liu Y, Zhang J et al (2017) Factors influencing soil enzyme activity in China’s forest ecosystems[J]. Plant Ecol 219(1):1–14
Zheng HF, Chen YM, Yang L et al (2018) Litter quality drives the differentiation of microbial communities in the litter horizon across an alpine treeline ecotone in the eastern Tibetan Plateau[J]. Sci Rep 8(1):10029
Zhong WH, Cai ZC (2004) Methods for studying soil microbial diversity. Chin J Appl Ecol 15(5):899–904
Acknowledgments
We would like to thank Shuai Yang, Jing Deng, Jiaqi Shi, Shiyu Tang, and other graduate students at the Institute of Ecology and Forestry, Sichuan Agricultural University, for assistance with field sampling and laboratory analyses.
Funding
This study was financially supported by the National Natural Science Foundation of China (31570605), the National key Research Program of China (2017YFC0505003), the key project of Sichuan education department (18ZA0393), and Key Research Program of Sichuan Province (18ZDYF0307).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Statement on ethical approval
Ethics Committee approval was obtained from the Institutional Ethics Committee of Miyaluo Nature Reserve to the commencement of this study.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Ana Rincón
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contribution of the co-authors
Yang Liu and Xian Shen designed the analysis; Yang Liu, Xian Shen, Yamei Chen, Lifeng Wang, and Qianmei Chen were involved in drafting or revising the manuscript. Yang Liu, Xian Shen, Yamei Chen, Lifeng Wang, Qianmei Chen, Jian Zhang, Zhenfeng Xu, Bo Tan, Li Zhang, Jiujin Xiao, Peng Zhu, and Lianghua Chen read and approved the final manuscript.
Annexes
Annexes
Rights and permissions
About this article
Cite this article
Liu, Y., Shen, X., Chen, Y. et al. Litter chemical quality strongly affects forest floor microbial groups and ecoenzymatic stoichiometry in the subalpine forest. Annals of Forest Science 76, 106 (2019). https://doi.org/10.1007/s13595-019-0890-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13595-019-0890-3