Abstract
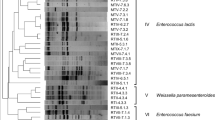
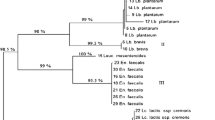
The microbiota of two traditional Iranian cheeses (Lighvan and Koozeh) made of raw ewe’s milk or mixtures of ewe’s and goat’s milk without starter addition was explored by culture-independent and culture-dependent approaches. Three batches of Lighvan and one of Koozeh were subjected to culture-independent polymerase chain reaction (PCR)–denaturing gradient gel electrophoresis (DGGE) analysis and sequencing of dominant bands to assess the microbial structure and dynamics through manufacturing and ripening. In addition, culturing in elective media for lactic acid bacteria (M17, MRS and KAA), isolation of single colonies (n = 130), molecular identification by PCR-amplified ribosomal DNA restriction analysis and sequencing, and differentiation at the strain level by repetitive extragenic palindromic PCR was also performed. DGGE analysis showed that the dominant amplicons in all four cheese batches belonged to Lactococcus lactis and Streptococcus parauberis. In addition, Escherichia coli and Lactococcus garvieae were frequently identified in both Lighvan and Koozeh, while Streptococcus thermophilus was found occasionally. In contrast, Enterococcus faecium and Enterococcus faecalis were found to be dominant among the isolates in all batches. These species showed a high genetic diversity. The discrepancy between culturing and DGGE results suggested that dominant populations were in a nonrecoverable state in the used media. This reinforces the idea that culture-dependent and culture-independent techniques provide complementary data, ultimately affording a better description of cheese ecosystems. These data could be of help in the selection of commercial starters for industrial-scale manufacture of Lighvan and Koozeh cheeses using pasteurised milk. Alternatively, microbial analysis would allow the selection of appropriate strains for designing of specific starters for traditional cheese manufacture.
伊朗传统Lighvan和Koozeh干酪的微生物多样性
摘要采用纯培养和非培养方法研究了由生鲜羊奶或者混合羊奶(羊奶和山羊奶)自然发酵制作的伊朗传统Lighvan和Koozeh干酪的微生物区系。对3批Lighvan干酪和1批Koozeh干酪进行了非培养的PCR-DGGE分析和主要条带的测序,以此评价干酪制作和成熟过程中微生物区系的结构和动力学。此外,采用选择性培养基(M17, MRS and KAA)对乳酸菌进行了培养,采用PCR-ARDRA、基因测序以及rep-PCR方法对分离出的单个菌落(n=130)从分子水平上进行鉴定。DGGE分析结果表明,在所有4批干酪样品中优势菌群为Lactococcus lactis a和Streptococcus parauberis。此外,在Lighvan和 Koozeh干酪中Escherichia coli和Lactococcus garviea的检出频率较高,但只在几个干酪样品中检测到Streptococcus thermophilus。相反,在所有样品中Enterococcus faecium 和Enterococcus faecalis也是主导菌群。微生物菌群之间表现出较高的生物多样性。纯培养和非培养的DGGE结果之间的差异表明主导菌群在这些培养基中是不可回收的,这种结果说明在进行干酪微生物生态系统的研究中,纯培养和非培养方法获得的数据可以互补。可以从获得的菌株中筛选出具有潜在工业化生产Lighvan和Koozeh干酪的发酵剂,也可以从中筛选出特定的菌株作为发酵剂用于传统干酪的生产。





Similar content being viewed by others
References
Abdi R, Shikh-Zeinoddin M, Soleimanian-Zad S (2006) Identification of lactic acid bacteria isolated from traditional Iranian Lighvan cheese. Pak J Biol Sci 9:99–103
Alegría A, Álvarez-Martín P, Sacristán N, Fernández E, Delgado S, Mayo B (2009) Diversity and evolution of the microbial populations during manufacture and ripening of Casín, a traditional Spanish, starter-free cheese made from cow’s milk. Int J Food Microbiol 136:44–51
Ayad EHE, Verheul A, Engels WJ, Wouters JT, Smit G (2001) Enhanced flavour formation by combination of selected lactococci from industrial and artisanal origin with focus on completion of a metabolic pathway. J Appl Microbiol 90:59–67
Ayad EHE, Verheul A, Wouters JTM, Smit G (2002) Antimicrobial-producing wild lactococci isolated from artisanal and non-dairy origins. Int Dairy J 12:145–150
Barouei J, Karbassi A, Ghoddusi HB, Mortazavi A (2008) Lactic microflora present in Liqvan ewe’s milk cheese. Int J Food Properties 11:407–414
BLAST (2011) Basic Local Alignment Search Tool, National Center for Biotechnology Information, National Library of Medicine (NLM), National Institutes of Health (NIH), USA. http://www.ncbi.nlm.nih.gov/BLAST/. Accessed Feb 2011
Cocolin L, Aggio D, Manzano M, Cantoni C, Comi G (2002) An application of PCR-DGGE analysis to profile the yeast populations in raw milk. Int Dairy J 12:407–411
Cocolin L, Diez A, Urso R, Ratnsiou K, Comi G, Bergmaier I, Beimfohr C (2007) Optimization of conditions for profiling bacterial populations in food by culture-independent methods. Int J Food Microbiol 120:100–109
Coppola S, Blaiotta G, Ercolini E, Moschetti G (2001) Molecular evaluation of microbial diversity in different types of Mozzarella cheese. J Appl Microbiol 90:414–420
El-Baradei G, Delacroix-Buchet A, Ogier JC (2007) Biodiversity of bacterial ecosystems in traditional Egyptian Domiati cheese. Appl Environ Microbiol 73:1248–1255
Ercolini D, Moschetti G, Blaiotta G, Coppola S (2001) The potential of a polyphasic PCR-DGGE approach in evaluating microbial diversity of natural whey cultures for water-buffalo Mozzarella cheese production: bias of culture-dependent and culture-independent analyses. Syst Appl Microbiol 24:610–617
Ercolini D, Hill PJ, Dood CER (2003) Bacterial community structure and location in Stilton cheese. Appl Environ Microbiol 69:3540–3548
Ercolini D, Mauriello G, Blaiotta G, Moschetti G, Coppola S (2004) PCR-DGGE fingerprints of microbial succession during a manufacture of traditional water buffalo mozzarella cheese. J Appl Microbiol 96:263–270
Fernández E, Alegría A, Delgado S, Mayo B (2010) Phenotypic, genetic and technological characterization of Lactococcus garvieae strains isolated from a raw milk cheese. Int Dairy J 20:142–148
Flórez AB, Mayo B (2006) Microbial diversity and succession during the manufacture and ripening of traditional, Spanish, blue-veined Cabrales cheese, as determined by PCR-DGGE. Int J Food Microbiol 110:165–171
Foulquié Moreno MR, Sarantinopoulos P, Tsakalidou E, de Vuyst L (2006) The role and application of enterococci in food and health. Int J Food Microbiol 106:1–24
Giraffa G (2003) Functionality of enterococci in dairy products. Int J Food Microbiol 88:215–222
Jany J-L, Barbier G (2008) Culture-independent methods for identifying microbial communities in cheese. Food Microbiol 25:839–848
Kafili T, Razavi SH, Emam Djomeh Z, Naghavi MR, Álvarez-Martín P, Mayo B (2009) Microbial characterization of Iranian traditional Lighvan cheese over manufacturing and ripening via culturing and PCR-DGGE analysis: identification and typing of dominant lactobacilli. Eur Food Res Technol 229:83–92
Koeuth T, Versalovic J, Lupski JR (1995) Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome Res 5:408–418
Lafarge V, Ogier J-C, Girard V, Maladen V, Leveau J-Y, Gruss A, Delacroix-Buchet A (2004) Raw cow milk bacterial population shifts attributable to refrigeration. Appl Environ Microbiol 70:5644–5650
Mayo B (2010) Dairy lactococci. In: Heldman RD, Bridges A, Hoover D, Wheeler M (eds) Encyclopedia of biotechnology in agriculture and food. Taylor & Francis Group, London. DOI: 10.1081/E-EBAF-120044696
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl Environ Microbiol 59:695–700
Ogier JC, Serror P (2008) Safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol 126:291–301
Ogier J-C, Lafarge V, Girard V, Rault A, Maladen V, Gruss A, Leveau J-Y, Delacroix-Buchet A (2004) Molecular fingerprint of dairy microbial ecosystems by use of temporal temperature denaturing gradient gel electrophoresis. Appl Environ Microbiol 70:5628–5643
Palys T, Nakamura LK, Cohan FM (1997) Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol 47:1145–1156
Parente E, Cogan TM (2004) Starter cultures: general aspects. In: Fox PO (ed) Cheese: chemistry, physics and microbiology, 3rd edn. Elsevier, Oxford, pp 123–147
Pitkälä A, Koort J, Björkroth J (2008) Identification and antimicrobial resistance of Streptococcus uberis and Streptococcus parauberis isolated from bovine milk samples. J Dairy Sci 91:4075–4081
Pogacic T, Samarjiza D, Coich V, D’Andrea M, Kagli DM, Giacomini A, Majhenic AC, Rogelj I (2010) Microbiota of Karakacanski skakutanak, an artisanal fresh sheep cheese studied by culture-independent PCR-ARDRA and PCR-DGGE. Dairy Sci Technol 90:461–468
Poznanski E, Cavazza A, Cappa F, Cocconcelli PS (2004) Indigenous raw milk microbiota influences the bacterial development in traditional cheese from an alpine natural park. Int J Food Microbiol 92:141–151
Randazzo CL, Torriani S, Akkermans ALD, de Vos WM, Vaughan EE (2002) Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl Environ Microbiol 68:1882–1892
Randazzo CL, Vaughan EE, Caggia C (2006) Artisanal and experimental Pecorino Siciliano cheese: microbial dynamics during manufacture by culturing and PCR-DGGE analyses. Int J Food Microbiol 109:1–8
Randazzo CL, Pitino I, De Luca S, Scifò GO, Caggia C (2008) Effect of wild strains used as starter cultures and adjunct cultures on the volatile compounds of the Pecorino Siciliano cheese. Int J Food Microbiol 122:269–278
RDP (2011) Ribosomal Database Project, Center for Microbial Ecology, Michigan State University, USA http://rdp.cme.msu.edu/index.jsp. Accessed Feb 2011
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Wouters JTM, Ayad EHE, Hugenholtz J, Smit G (2002) Microbes from raw milk for fermented dairy products. Int Dairy J 12:91–109
Acknowledgements
This research was partially supported by a project from the Spanish Ministry of Science and Innovation (MICINN) to BM (Ref. AGL2007-61869-ALI). AA was awarded a scholarship of the Severo Ochoa programme from FICYT (Ref. BP08-053). The authors wish to thank the Iranian Ministry of Industries and Mines, as well as Razavi Dairy Industry (Mashhad, Iran) and the Office of Industrial Relationships (OIR) of Ferdowsi University of Mashhad (FUM).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Microbial dynamics as determined by identification of DGGE bands through manufacture and ripening stages of the traditional Iranian cheeses Lighvan and Koozeh (DOC 93 kb)
Online Resource Fig. 1
Partial amplified ribosomal DNA restriction analysis (ARDRA) profiles of 11 species from Lighvan and Koozeh cheeses from different manufacturing and ripening stages. The 16S rRNA gene was amplified using primers 27F and 1492R and digested with the restriction enzymes HaeIII (a) and HhaI (b). M molecular weight marker GeneRuler™. After partial amplification, sequencing and sequence comparison of 16S rRNA genes, the profiles were shown to correspond to the following species: 1 E. faecium; 2 L. plantarum, 3 L. brevis, 4 L. lactis subsp. lactis, 5 E. faecalis, 6 Enterococcus durans, 7 Enterococcus casseliflavus, 8 Enterococcus italicus, 9 Micrococcus luteus; 10 S. haemolyticus, and 11 Aerococcus viridans (PPT 369 kb)
Online Resource Fig. 2
Repeatability of the REP-PCR typing assay with primer BoxA2R after analysing of three randomly selected isolates (a, b, and c) in three independent experiments (1, 2, and 3). Below, dendogram of similarity of the different typing patterns clustered by the UPGMA method using the Simple Matching coefficient. M GeneRuler™ (PPT 163 kb)
Online resource Fig. 3
Typing REP-PCR profiles obtained with primer BoxA2R among the 36 E. faecium isolates from Koozeh cheese. Below, dendogram of similarity of the different typing patterns clustered by the UPGMA method using the Simple Matching coefficient. M GeneRulerTM. The broken line denotes the arbitrary percentage of similarity used to consider isolates as different strains (PPT 895 kb)
About this article
Cite this article
Edalatian, M.R., Najafi, M.B.H., Mortazavi, S.A. et al. Microbial diversity of the traditional Iranian cheeses Lighvan and Koozeh, as revealed by polyphasic culturing and culture-independent approaches. Dairy Science & Technol. 92, 75–90 (2012). https://doi.org/10.1007/s13594-011-0045-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13594-011-0045-2