Abstract
The recycling of organic waste worldwide is not effective, which leads to water pollution and loss of potential crop fertilizers. Available resources have to be used more efficiently as the world population increases. An innovative solution is to use insects for the management of organic waste. Here, we used black soldier fly to convert organic waste into animal feed protein, as fly larvae, and plant fertilizer, as compost residue. A continuous fly reactor was monitored for 9 weeks. We analyzed physicochemical and microbial parameters, and we evaluated the sanitary risk. Results show 55.1 % of material degradation and 11.8 % of biomass conversion based upon total solids. We observed higher levels of N and P in the treatment residue than in the inflow material. Results also show a lower concentration of Salmonella spp. and viruses. Compost treatment with black soldier fly is therefore an efficient system for nutrient recycling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The volume of solid waste generated every year is increasing worldwide. Although municipalities strive to provide solid waste management, much of the generated waste is left untreated (Hoornweg and Bhada-Tata 2012). A large fraction of solid waste in low- and middle-income countries consists of organic material, e.g., food waste, animal manure, and market waste. If left untreated, the valuable energy and nutrients contained in the waste can contribute to an increase in annual greenhouse gas emissions (UNEP 2010) and a leaching of nutrients into water bodies, causing eutrophication (Sharpley et al. 1994). Over and above this, there is a risk of spreading the pathogens contained in the organic waste.
The waste management coverage in high-income countries is considerably higher, but at the same time the waste generated per person is up to three times greater and still increasing (Hoornweg and Bhada-Tata 2012). The proportion of organic waste is around 30 % and only a small proportion of the nutrients returns into the food cycle (Cordell et al. 2009). This is a growing concern, since the nitrogen and phosphate cycle has been identified as key factors that has to be maintained within certain levels for the planet to be able to support human existence in the future (Rockström et al. 2009). One way of securing human well-being is, according to Griggs et al. (2013), sustainable production systems. In a report by the United Nations Food and Agricultural Organization, it was stated that the consumption of insects in the food sector, as animal feed and directly for human consumption, is one such solution (van Huis et al. 2013).
The black soldier fly, encountered between 46°N and 42°S (Martínez-Sánchez et al. 2011), is of particular interest. Their ability to consume prodigious amounts of organic waste has long been known. Black soldier fly larvae have been reported to consume and degrade a number of organic materials with material degradation up to 70 % (Diener et al. 2011). The larvae in the final larval stage, the prepupae, crawl out of the feeding material to find a dry and dark place to pupate (Sheppard et al. 2002) and are thus self-harvesting.
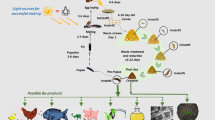
The prepupae contain around 40 % protein and 30 % fat and have been demonstrated to be a good alternative feed in fish (St-Hilaire et al. 2007) and pig (Newton et al. 1977) production, with the potential to replace fish meal and fish oil as source of animal feed protein. Replacing fish meal would alleviate the pressure on the wild fish population (Tacon and Metian 2008). With the increasing consumption of fish and meat, the demand for animal feed protein has increased greatly, while its availability is limited. As a consequence, the price of fish meal has reached new heights -- valued USD 1,820 per tons (World Bank, May 2014) -- making the production of compatible alternative animal feed protein a lucrative business. By producing an economically valuable product, organic waste management could become a financially sustainable system that would increase resource reuse efficiency and reduce negative environmental impacts (Fig. 1).
A graphical representation of the concept of fly larvae composting: organic waste is consumed by fly larvae, in the sixths and final larval stage, the prepupae migrate out of the compost. The prepupae can be used as animal feed and the treatment residue used as organic fertilizer. The loop is closed when the animal manure and food waste is diverted as substrate into the fly larvae compost
Hygiene aspects play an important role when dealing with organic waste and animal feed (Sidhu and Toze 2009). In several studies, black solider fly larvae waste treatment systems have been shown to remove bacteria in the Enterobacteriaceae family: the concentration of Salmonella spp. was reduced by 7 log10 in 8 days in a small batch-operated fly larvae system treating fecal sludge (Lalander et al. 2013); Erickson et al. (2004) observed the reduction of Salmonella enterica Serovar Enteritidis and Escherichia coli O157:H7 in cattle manure, while Liu et al. (2008) have reported on the reduction of E. coli in poultry manure. However, although a high reduction in bacteria of the Enterobacteriaceae family occur, reduction of other organisms has not been observed. All these systems were operated in batch mode. The question is whether the same results would be obtained in a continuous flow reactor. In a large-scale fly larvae waste management system, it is likely that a continuous flow reactor would be implemented, as it would be too time-consuming to operate in batch mode. One hypothesis is that the operation over a long period would support the buildup of a microbial community, helping to break down the waste material and thus improve the hygienic quality of the treatment residue.
The objective of this study was to establish a continuous flow fly larvae reactor and to analyze important process and hygiene parameters in order to evaluate material degradation, waste-to-biomass conversion rate, nutrient flow, and average weekly reduction of evaluated pathogens and indicator organisms.
2 Materials and methods
2.1 Materials
Pig manure (21.1 ± 1.7 % total solids) was collected on a pig farm in Uppsala municipality, Sweden. Dog food (Puppy Original, Pro Plan, Purina) mixed with water (21.9 ± 0.2 % total solids) was used as the model substrate for organic waste (Vinnerås et al. 2003). Human feces were collected fresh in plastic bags and stored at −20 °C immediately upon collection. A mixture of pig manure, dog food, and human feces was prepared (4:4:2; 28.7 ± 1.2 % total solids), divided into feeding portions, and kept at −20 °C until use.
Ascaris suum, which infects pigs, is often used as model for Ascaris lumbricoides, which infects humans (Johnson et al. 1998). A. suum extracted from the feces of infected pigs were purchased from Excelsior Entinel, Inc. and received in aqueous solution at a concentration of 100,000 eggs mL−1.
The bacterial inoculate solutions consisted of 108 colony forming units (CFU) mL−1 Enterococcus faecalis (ATCC 29212) and 109 CFU mL−1 Salmonella enterica subspecies ~ 1 serovar Typhimurium phage type 178 (isolated from sewage sludge (Sahlström et al. 2004), grown in unselective bacterial nutrient medium (NB, Oxoid AB, Sweden).
Propagation of bacteriophage фX174 to a concentration of 109 plaque forming units (PFU) mL−1 was performed in unselective bacterial nutrient medium (NB, Oxoid AB, Sweden) using the host strain Escherichia coli (ATCC 13706). The phage was collected by centrifuging the solution at 2,000 × g for 10 min followed by sterile filtration and kept at 4–6 °C until use.
The virus inoculate comprised the following: Reovirus type 3 (ATCC VR-232); strain Abney (human); genus Orthoreovirus; Canine Adenovirus I, strain vacc-98, genus Mastadenovirus; and Porcine enterovirus I, genus Teschovirus.
Black soldier fly eggs were received twice a week from the Research Institute of Organic Agriculture, Frick, Switzerland.
2.2 Experimental set-up and sampling
Three fly larvae reactors consistent of white 37 L Sortera bins (37 × 55 × 28 cm), with a lid and slanting wall for prepupal migration, that were placed inside 45 L black Samla bins (39 × 57 × 28 cm) with a lid, both from IKEA (Uppsala, Sweden). The Samla bins were used to keep light out of the reactor and as collection bins for migrating prepupae. The reactors were kept at 25 °C throughout the experiment.
Black soldier fly egg packages were received in cardboard strips. Individual egg packages were divided into small (around 150–250 larvae), medium (around 400–500 larvae), and large (around 800–1,000 larvae) egg packages and hatched individually. Twice a week, a more or less equal number (based on the number and size of egg packages) of 5–10-day-old larvae were added to each reactor. The eggs were hatched in the same feed mixture used in the reactors. The total number of larvae at steady-state was estimated to be 7,200.
The reactors were fed three times a week. The pre-prepared feed mixture was thawed overnight and added in the mornings. The feed portions were adjusted to the estimated number of larvae present in the reactors, to a feeding rate of 100 mg per larvae per day (Diener et al. 2009); at steady-state each unit was fed 5.4 kg per week.
The reactors were operated as plug-flow: old material in the back of the reactor was pushed 5 cm to the front prior to fresh feed mixture being added. All samples were collected from the front 10 cm of the reactor.
On each feeding occasion, the feed mixture was inoculated with Salmonella typhimurium (1 % w/w), E. faecalis (1 % w/w), and фX174 (1 % w/w).
On the first feeding occasion in week 4, the feed mixture was inoculated with 1 mL A. suum of concentration 100,000 egg mL−1.
On the first feeding occasion in week 7, the feed mixture was inoculated with Reovirus type 3, Canine Adenovirus I, and Porcine enterovirus I.
The reactors were weighed weekly throughout the course of the experiment. All migrating prepupae were counted/weighed. Once a week, triplicate samples of 5 g were collected in each reactor and analyzed separately to determine the concentration of bacteria and bacteriophages in the influent and effluent material. The concentration and viability of A. suum eggs was determined on days 7, 14, and 28 after inoculation. The concentration of viruses in the material was determined 2, 4, 7, 9, 11, and 14 days after inoculation. Samples for physicochemical analysis were collected at the start and end of the experiment.
2.3 Physicochemical analyses
The material was dried at 105 °C for 14 h to determine total solids and at 550 °C for 6 h to determine volatile solids, i.e., the organic content.
A radiometer electrode was used to measure pH. All analyses were conducted at room temperature on 10 g of sample diluted with 50 mL deionized water and were left to settle for 1 h prior to analysis.
In a closed container, 1 g of sample was mixed with 20 mL deionized water. The material was thoroughly mixed and diluted 1:10 in deionized water. Spectroquant© test kit number 114544 was used for total ammonium nitrogen (NH4-N) analyses and kit number 114764 for nitrate (NO3-N) analyses.
In a 50-mL volumetric flask, 0.5–0.6 g samples were dissolved into 10 mL concentrated H2SO4 and brought to “rolling boil” on a hotplate. When the sample had visually dissolved, an additional 5 mL H2SO4 was added. The sample was allowed to cool and was thereby diluted—1:100 for total nitrogen analysis and 1:50 for total phosphorous analysis—in deionized water. Spectroquant© test kit number 1.00613.0001 was used for total nitrogen analysis and number 1.00673.0001 was used for total phosphorous analysis.
2.4 Microbiological analyses
2.4.1 Bacterial and bacteriophage analyses
The buffer used for all experiments was buffered NaCl peptone water with Tween 80 at pH 7, hereafter referred to as Tween buffer. Sample (5 g) was extracted and further diluted in the Tween buffer.
A 100 µL volume of selected dilution was spread on Slanetz–Bartley agar (Oxoid AB, Sweden) and incubated at 44 °C for 48 h for Ent. spp. enumeration and on xylose lysine desoxycholate agar (XLD) (Oxoid AB, Sweden) containing 0.15 % sodium-novobiocin and incubated at 37 °C for 12 h for Salmonella spp. enumeration. The plates were enumerated with a detection limit of 100 CFU mL−1.
To lower the detection limit of Salmonella spp. concentration, 200 μL per plate of sample of dilution 10−1 was spread on five plates per sample and enumerated with a detection limit of 10 CFU mL−1. To lower the detection limit further, the sample was enriched as follows: 5 g per unit was immersed in 45 mL buffered peptone water and incubated at 37 °C for 24 h, upon which 100 μL was immersed into Rappaport–Vassiliadis salmonella enrichment broth (RVB) and incubated at 41.5 °C for 24 h. Positive results were evaluated by spreading 10 μL on a XLD plate and incubated at 37 °C for 12 h, lowering the detection limit to 1 CFU g−1.
Total thermotolerant coliforms were enumerated in double layer agar using violet red bile agar (VRB) (Oxoid AB, Sweden); 1 mL of sample was mixed with 7–8 mL of agar, and upon solidification of the first layer, an additional 7–8 mL agar was added. The plates were incubated at 44 °C for 24 h and counted with a detection limit of 10 CFU g−1.
For sampling of coliphage фX174, the host E. coli (ATCC 13706) was cultured in unselective microbial medium (NB, Oxoid AB, Sweden) at 37 °C for up to 4 h. A 1 mL sample volume of suitable dilution was mixed with 2 mL soft agar and 1 mL host solution, poured onto blood agar base (BAB) plates (Oxoid AB, Sweden) and incubated at 37 °C for 16 ± 2 h.
2.4.2 A. suum ova extraction
A. suum ova extraction was conducted according to the procedure of the United States Environmental Protection Agency (EPA/625/R-92/013), using 30 g of material. Prior to incubation, 15–20 of the extracted ascaris eggs were verified. The tubes were incubated at 28 °C for 30 days in 0.1 N H2SO4.
After approximately 30 days, the incubated material was analyzed under the microscope (~100 ova per sample). A 1 mL Sedgwick Rafter Counting Cell was used to count eggs; pre-larvae and larvae were counted as viable.
2.4.3 Virus analyses
Samples for virus analysis were diluted 1:5 in NaCl peptone water with Tween buffer, further diluted 1:2 in beef extract with glycine, and kept at −70 °C until analysis. After thawing, the samples were vortexed for 15 min at 60 rpm, centrifuged at 3,000 × g for 15 min at 5 °C, and the supernatant filtered through a 0.45 μm filter. The samples were then filtered through a sterile gel filter (PD10, GE Healthcare) to remove toxicity. The samples were analyzed by endpoint titration using the following cell cultures: MDCK (ATCC CCL-34) for adenovirus, BHK (ATCC CCL-10) for reovirus, and swine kidney cell line (SK-6) for enterovirus, all cells were prepared 24 h prior to infection. To assess the virus concentration in the ingoing material, the samples were diluted tenfold and eight replicates of 50 μL were assayed for each dilution, while 48 replicates of the lowest dilutions were used to lower the detection limit.
2.5 Calculation
2.5.1 Material conversion
The waste-to-biomass conversion rate (BCR) on a total solids basis was calculated by:
where Mig.PPTot.TS and WasteTot.TS were the total solids in the migrating prepupae and waste, respectively.
The calculations for the nutrient flow balance and material degradation were based on the assumption that:
where WasteTot.Ash, ResidueTot.Ash, and Mig.PPTot.Ash were the total amount of ash in the inflow waste, the residue, and the migrated prepupae, respectively. The amount of ash in the residue was estimated based on the measured values in the inflow material and migrated prepupae.
The material reduction (MatTS.Red) on a total solids basis was calculated as:
where ResidueTot.TS and WasteTot.TS were the total amount of total solids in the treatment residue and the inflow waste, respectively.
2.5.2 Reduction of microorganisms
The log10 reduction of evaluated microorganisms that occurred during 1 week in the treatment (∆ log10 Red) was calculated as:
where \( {C}_{{\mathrm{Mat}}_i} \) was the estimated concentration of the material in the treatment unit after substrate inoculation week i and \( {C}_{\mathrm{Mat}.{\mathrm{out}}_{i+1}} \) the theoretical concentration of the material in the treatment unit prior to substrate inoculation 1 week later (i + 1).
The theoretical concentration of the material in the treatment unit week i (\( {C}_{{\mathrm{Mat}}_i} \)) was calculated as:
where \( {C}_{\mathrm{Sub}.{\mathrm{in}}_i} \) and \( {m}_{\mathrm{Sub}.{\mathrm{in}}_i} \) were the concentration and mass, respectively, of the inflow substrate week i and \( {C}_{{\mathrm{Mat}}_i} \) and \( {m}_{{\mathrm{Mat}}_i} \) the concentration and the mass, respectively, of the treatment unit week i.
2.5.3 Mass balance
The material in the fly larvae composts was assumed to be mainly polysaccharides (cellulose and starch); thus, the chemical formula C6H10O5 was used when calculating the amount of oxygen needed for the respiration. For each polysaccharide, six oxygen molecules were required for full degradation into six carbon dioxide and five water molecules. For the mass balance, it was assumed that the total mass of material reduced on organic content basis had been completely respired into carbon dioxide and water.
The nutrient mass balance was conducted by multiplying the measured concentration of the evaluated nutrients with the mass of the total solids, for each fraction.
2.6 Statistical analyses
Two-tailed, paired Student’s t test (95 % confidence interval) was used to evaluate whether the reduction of microorganisms was significant and if a statistically significant difference occurred between the total solids, organic content and the concentrations of nutrients in the inflow and outflow materials. Linear regression with t test was used to evaluate if the pH and the reduction of microorganisms changed over time. All analyses and graphical presentations were conducted in R (R Core Team 2012).
3 Results and discussions
Three black soldier fly compost reactors were continuously fed a mixture of human and pig manure and organic waste during 9 weeks. Once weekly, samples of the outflow material were collected and physicochemical and microbial parameters analyzed. The process efficiency in the fly larvae reactor was verified by determining the material degradation, waste-to-biomass conversion rate, mass balance of the organic content and nitrogen and phosphorous, the average weekly reduction of the evaluated microorganisms, and the survival of Adeno-, Reo- and Enterovirus and A. suum eggs in the process.
3.1 Change in physicochemical properties
The organic content significantly (p < 0.05) decreased in the process, from 86.6 % in the inflow material to 72.9 % in the outflow material (Table 1). The pH was found to significantly (p < 0.05) increase with time; the average pH in the inflow material was 5.6, while the average pH in the outflow material was 8.3. There was no significant (p > 0.05) difference in the change in pH over time between the three reactors. The prepupae were 12.3 % ash and 59.9 % moisture.
The concentration of total phosphorous significantly (p < 0.05) increased per gram total solids in the process, from around 33 mg g−1 to almost 48 mg g−1 (Table 2). The concentration of total nitrogen, organic nitrogen and nitrate was not significantly different (p < 0.05) in the inflow and outflow material, while the concentration of total ammonium nitrogen significantly increased (p < 0.05). Some of the nitrogen was incorporated into the prepupal biomass in the form of proteins. The nitrogen profile changed significantly in the process: the organic nitrogen present in the inflow material was converted into ammonia, changing the organic nitrogen to total nitrogen ratio from 0.91 to 0.78 and the C/N ratio decreased from 16.4 to 14.3.
3.2 Material conversion and nutrient flows
The total flow of nutrients was estimated for the fly larvae composting process (Fig. 2). It was found that close to half of the total nitrogen in the inflow material was degassed as ammonia, while just over 10 % was incorporated into the larval biomass. Approximately 30 % of the total phosphorous and 10 % of the ash was not accounted for in the system, as these two components should not disappear it is likely that it is due to experimental and/or analytical factors. The flow balance was estimated by multiplying the concentration of total phosphorous for the inflow and outflow with the corresponding mass. It could be that the total phosphorous concentration was not constant for all outflow material. Less than 3.5 % of the total phosphorous was incorporated into the larval biomass.
The total material reduction was 55.1 % on a total solids basis. Of the total 32 kg flowing into the fly larvae compost, around 20 kg turned into carbon dioxide and water and 2.4 kg (11.8 %) was converted into larval biomass (Table 3). The concentration of total ammonium nitrogen increased, while the concentration of organic nitrogen decreased by almost 63 %.
The waste-to-biomass conversion rate of 11.8 % on a total solids basis presented in this paper only included the migrating prepupae. Sheppard et al. (1994) reported an optimal conversion rate of 8 % on a total solids basis in a low-maintenance system operating in ambient temperature, converting hen manure into prepupal biomass. The optimal conversion rate was found for the May to August period, while a considerably lower conversion rate was found for the period May to December. In good accordance with this study, Diener et al. (2011) reported a conversion rate of 11.8 % on a total solid basis in mixed organic waste. The highest waste-to-biomass conversion rate for black soldier fly larvae reported hitherto is 22.9 %, in which human feces was converted to prepupal biomass in a small-scale system feeding ten larvae one lump sum of material (Banks et al. 2014). The conversion rate drastically decreased to 1.6 % in a treatment with 100 larvae with lump sum feeding, while it was 15.9 % with incremental feeding with the same number of larvae. It appears that several factors impact the conversion rate: the waste source is a major contributor, but also the temperature at which the system operates, the larval density and the feeding regime appears to have an impact.
In this study, it was observed that some prepupae never left the reactor, but pupated in the outflow region. A higher waste-to-biomass conversion rate could therefore be expected in a more optimized system. One way of forcing the prepupae to leave the system could be by increasing the moisture percentage in the outflow region. Moreover, the pig manure fraction contained a considerable proportion of straw, which cannot be consumed by the larvae. A substrate with lower proportion of straw would also yield a higher conversion rate.
The material degradation was over 50 %, in accordance with previous findings for degradation of different animal manures (Myers et al. 2008; Sheppard et al. 1994). For other feed stuff, higher degradation has been achieved. A 70 % degradation on a total solids basis was demonstrated for mixed organic waste (Diener et al. 2011) and hen feed (Gobbi et al. 2013), while a 75 % reduction on a total solids basis was demonstrated in fecal sludge (Lalander et al. 2013). The higher degradations observed in those materials is probably due to a higher percentage of easily available carbon. The straw in the pig manure made the waste substrate less degradable to the larvae.
The total phosphorous was found to be concentrated in the outflow material, which decreases the amount of field work during application.
Around 44 % of the total nitrogen evaporated, mainly as ammonia. Ammonia evaporation is governed by the ammonium–ammonia equilibrium, which in turn depend on the pH and temperature (Emerson et al. 1975). This system was run at 25 °C and the pH increased from pH 5.6 at the start to pH 8 after 3 weeks of treatment (Table 1). As a consequence, the evaporation of ammonia can become considerable in this type of well-ventilated system. In a large-scale system with controlled ventilation, emissions of ammonia can be kept low by condensation of the outgoing air. Preliminary studies on batch fly larvae systems demonstrated that the emissions of nitrous oxide (N2O) were so small as to be negligible (unpublished data). Of the total amount of nitrogen in the system, loss through denitrification is likely to be small since the amount of nitrate in the system was not great and the process was relatively well-aerated through larval movement. Although a considerable proportion of the total nitrogen was lost, the concentration of total ammonium nitrogen was higher in the outflow than in the inflow, thereby improving the fertilizer value of the treatment residual in comparison to untreated material.
The outflow material that may be used as organic fertilizer comprised on a total solids basis 4.8 ± 0.10 % total phosphorous and 5.1 ± 0.2 % total nitrogen, of which around 77.9 ± 3.0 % was organic nitrogen and 19.4 ± 2.3 % total ammonium nitrogen -- 0.98 ± 0.15 % of total residue (Table 2).
3.3 Hygienic quality of treatment residue
The concentration of Salmonella spp. reduced drastically in the process, from an average inflow concentration of around 107 CFU g−1 to below the detection limit of 1 CFU g−1. The concentration in the inflow was not measured in weeks 5–7, but is assumed to be similar to the concentrations measured in weeks 3–5 (Figs. 2 and 3a). The concentration of total thermotolerant coliforms appeared to decrease in the material (Figs. 2 and 3b), while the concentration of Enterococcus spp. did not appear to be reduced in the process (Figs. 2 and 3c). The inflow concentration of bacteriophage фX174 varied somewhat, but the concentration in the outflow was quite stable from week 5 onwards (Figs. 2 and 3d).
In order to get an estimation of the reduction of the evaluated organisms in the treatment, the average log10 reduction that occurred during one week in the treatment was calculated (5). In average, total thermotolarent coliforms and фX174 was reduced 0.6 and 1.7 log10, respectively, in one week (Table 4). The weekly reduction of thermotolerant coliforms was found to increase with time. No statistically significant improvement in weekly reduction of the фX174 was found over time. No reduction of Enterococcus spp. occurred in the treatment. There were no significant difference in the reduction of microorganisms between the reactors (p > 0.05).
One of the hypotheses in this study was that a continuous system could improve the hygienic quality of the fly larvae-treated material compared to a batch system, due to interactions with an improved microbial and fungal community. For the bacteria and bacteriophages, the results agreed with the findings for a batch system, with no improved reduction (Lalander et al. 2013). In the previous study, the reduction of bacteriophages was found to be equal in the larvae treated and control material and was thus concluded not to be affected by the larval activity. The weekly log10 reduction of thermotolerant coliforms was found to be 0.6 log10 in this experiment, but was found to improve with time. No identification of the specific coliform species was performed; however, it is likely that a large part of the coliforms included in the analyses were not E. coli. Erickson et al. (2004) demonstrated a greater reduction in E. coli O157:H7 at 27–31 °C in fly larvae-treated chicken manure compared to the reduction found in cattle manure. The reduction was increased with increasing temperature and similar reduction as observed in this study was observed in chicken manure at 23 °C. Chicken manure is considerably higher in ammonia than cattle manure (Paul and Beauchamp 1989); uncharged ammonia is well known to be toxic to microorganisms and as the concentration of uncharged ammonia increases with temperature (Nordin et al. 2009). The reduction of E. coli observed by Erickson et al. (2004) in chicken manure and the increased reduction observed at elevated temperatures could therefore have been caused by increased concentration of uncharged ammonia and be unrelated to larval activity.
The mechanism behind the reduction of salmonella observed in this study is not known. Choi et al. (2012) found that methanol extracts of the black soldier larvae had antibacterial effects on the Gram-negative bacteria studied, but no such effect was seen on the Gram-positive ones. The authors speculated that the antibacterial effect could be due to interaction between the bacterial ribosome or bacterial cell wall and the active substance. In accordance with Choi et al. (2012), a reduction was seen for the Gram-negative salmonella and some reduction for thermotolerant coliforms in this study, while no reduction of the Gram-positive enterococcus occurred.
The concentration of viable viruses decreased in the material, and was below the detection limit in all samples after 14 days (Table 5). Adenovirus was the most persistent of the viruses studied, while enterovirus was less persistent than reovirus.
The maximum virus concentrations in the material, which were recorded at day 2 for all viruses, were 2.5–4 log10 units below inflow concentration. This is more than the approximate 1 log10 dilution of the ingoing material—assuming that the inflow material was mixed evenly in the compost—and is probably due to inactivation of the viruses, possibly in combination with the non-uniform distribution of viruses in the material. As the virus concentration in the outgoing material decrease with time, an increased time interval between feeding and harvesting will yield a lower virus concentration in the outgoing material. There are several factors that affect the survival of viruses: larval activity can be one; a natural time-dependent die-off in ambient temperature another; adsorption onto particles is a third; and ammonia inactivation can be a fourth effect. Ammonia has been proven to have a good reduction effect on single stranded viruses (Emmoth et al. 2011). A study on the inactivation of adenovirus (ad41) during storage reported a decimal reduction time of 12.5 days at 20 °C in biosolids which have similar dry matter content (Wei et al. 2009). This is considerably longer than the findings in this study. Further investigations are required for a better understanding of the mechanisms involved.
The concentration and viability of ascaris eggs did not change significantly (p > 0.05) in the course of the treatment, nor was there any significant difference in concentration and viability of ascaris eggs between the three units over the course of the experiment (p > 0.05). The viability of the eggs was 90.3 % when they were inoculated into the treatment, 82.9 ± 4.3 after 1 week and 78.6 ± 6.7 after one month in the system. The concentration was 72.6 ± 11.7 eggs per gram after one week and 84.6 ± 68.8 after one month.
As for the bacteria and bacteriophages, the reduction of Ascaris eggs did not appear to change in the continuous system compared to what was found for the batch system (Lalander et al. 2013). It was not possible to determine the total reduction/destruction of ascaris eggs. When attempting to estimate the flow of the eggs in the system, based on the concentrations found in the outflow material, the total number of eggs exceeded the total number added, although no eggs were found in the fresh inflow material. Due to the adhesive nature of the ascaris eggs, it is likely that the concentration of eggs was not constant in the system. In this system, not much material was removed throughout the experiment due to the slow accumulation of solids during the 10 weeks of the study. Thus, it is possible that the eggs accumulated in the outflow region while the concentration of eggs could have been lower in the inflow region. However, as the concentration of eggs did not decrease with time in the outflow region, it does not appear that the eggs were destroyed. The viability of the eggs was not significantly (p > 0.05) reduced with time. However, it was observed that the eggs extracted after one month in the system had not started to develop into prelarvae after 30 days in the fly larvae reactor at 25 °C, which could otherwise be expected. After 30 days at 28 °C after the eggs were separated from the material, 79 % of the eggs had developed into prelarvae. It appeared that the development of the eggs had been inhibited during the treatment, despite the temperature being in a range that supports development. One reason could be the increased total ammonium nitrogen concentration, as uncharged ammonia is known to inactivate the development of ascaris eggs (Fidjeland et al. 2013). Additionally, Fidjeland et al. (2013) found that the main effect of ammonia was to shorten the lag phase for the bilinear inactivation of ascaris. It is therefore possible that a longer retention time in the fly larvae system or during post-storage could lead to inactivation of the ascaris eggs present.
3.4 Sanitary risks of the fly larvae system
The minimum retention time of material in the fly larvae compost was set at two days, while the hydraulic retention time was over 30 days. This was enough for inactivation of the zoonotic salmonella bacteria, while the other investigated organisms survived the treatment. Viruses had an indicated reduction of 2 log10 whereas ascaris eggs were not inactivated/destroyed. System management is therefore required that results in additional security in terms of breaking disease transmission, e.g., the selection of crops to be fertilized and incorporation of the fertilizer into the soil.
In a previous study, it was shown that the content of the zoonotic bacteria such as salmonella was reduced in the material to such a degree that it was not detectable in the harvested prepupae (Lalander et al. 2013). Other zoonotic bacteria were not tested, but in other studies a, similar reduction of another Gram-positive bacteria, E. coli O157, has been observed (Erickson et al. 2004). Due to the reduction of zoonotic bacteria, the risk of disease transmission -- when feeding animals other than the ones providing the feed to the larvae -- is a low-risk management system. For preservation and greater hygienic security, it may be required to process the prepupae further. With drying for example, the low water content and the temperature required for drying improve the hygienic status of the material, while preserving the larval meal at the same time.
4 Conclusions
The material degradation was 55.1 % and the waste-to-biomass conversion rate 11.8 % on a total solids basis in the continuous fly larvae composting system. Optimizing prepupal migration efficiency could increase the waste-to-biomass conversion rate. A large proportion of the nitrogen evaporated as ammonia: these losses can be kept low by condensing the outgoing air. Both the phosphorous and total ammonium nitrogen concentrations increased in the outflow material. The hygienic quality of the material was found to increase in the fly larvae treatment: the pathogen Salmonella spp. and viruses were reduced in the system, while the reduction of the other organisms studied was small or insignificant. Minimal retention time was found to be a key factor for pathogen reduction.
References
Banks IJ, Gibson WT, Cameron MM (2014) Growth rates of black soldier fly larvae fed on fresh human faeces and their implication for improving sanitation. Tropical Med Int Health 19(1):14–22. doi:10.1111/tmi.12228
Choi W-H, Yun J-H, Chu J-P, Chu K-B (2012) Antibacterial effect of extracts of Hermetia illucens (Diptera: Stratiomyidae) larvae against Gram-negative bacteria. Entomol Res 42(5):219–226. doi:10.1111/j.1748-5967.2012.00465.x
Cordell D, Drangert J-O, White S (2009) The story of phosphorus: global food security and food for thought. Glob Environ Chang 19(2):292–305. doi:10.1016/j.gloenvcha.2008.10.009
Diener S, Zurbrugg C, Tockner K (2009) Conversion of organic material by black soldier fly larvae: establishing optimal feeding rates. Waste Manag Res 27(6):603–610
Diener S, Studt Solano N, Roa Gutiérrez F, Zurbrügg C, Tockner K (2011) Biological treatment of municipal organic waste using black soldier fly larvae. Waste Biomass Valorization 2(4):357–363. doi:10.1007/s12649-011-9079-1
Emerson K, Russo RC, Lund RE, Thurston RV (1975) Aqueous ammonia equilibrium calculations: effect of pH and temperature. J Fish Res Board Can 32(12):2379–2383. doi:10.1139/f75-274
Emmoth E, Ottoson J, Albihn A, Belak S, Vinnerås B (2011) Ammonia disinfection of hatchery waste for elimination of single-stranded RNA viruses. Appl Environ Microbiol 77(12):3960–3966
Erickson MC, Islam M, Sheppard C, Liao J, Doyle MP (2004) Reduction of Escherichia coli O157:H7 and Salmonella enterica serovar enteritidis in chicken manure by larvae of the black soldier fly. J Food Prot 67(4):685–690
Fidjeland J, Magri ME, Jönsson H, Albihn A, Vinnerås B (2013) The potential for self-sanitisation of faecal sludge by intrinsic ammonia. Water Res 47(16): 6014–6023 doi:10.1016/j.watres.2013.07.024
Gobbi P, Martinez-Sanchez A, Rojo S (2013) The effects of larval diet on adult life-history traits of the black soldier fly, Hermetia illucens (Diptera: Stratiomyidae). Eur J Entomol 110(3):461–468
Griggs D, Stafford-Smith M, Gaffney O, Rockstrom J, Ohman MC, Shyamsundar P, Steffen W, Glaser G, Kanie N, Noble I (2013) Sustainable development goals for people and planet. Nature 495(7441):305–307. doi:10.1038/495305a
Hoornweg D, Bhada-Tata P (2012) What a waste—a global review of solid waste management. Urban Development Series. World Bank, Washington DC
Johnson PW, Dixon R, Ross AD (1998) An in-vitro test for assessing the viability of Ascaris suum eggs exposed to various sewage treatment processes. Int J Parasitol 28(4):627–633
Lalander C, Diener S, Magri ME, Zurbrügg C, Lindström A, Vinnerås B (2013) Faecal sludge management with the larvae of the black soldier fly (Hermetia illucens)—from a hygiene aspect. Sci Total Environ 458–460:312–318. doi:10.1016/j.scitotenv.2013.04.033
Liu Q, Tomberlin JK, Brady JA, Sanford MR, Yu Z (2008) Black soldier fly (Diptera: Stratiomyidae) larvae reduce Escherichia coli in dairy manure. Environ Entomol 37(6):1525–1530. doi:10.1603/0046-225X-37.6.1525
Martínez-Sánchez A, Magaña C, Saloña M, Rojo S (2011) First record of Hermetia illucens (Diptera: Stratiomyidae) on human corpses in Iberian Peninsula. Forensic Sci Int 206(1–3):e76–e78. doi:10.1016/j.forsciint.2010.10.021
Myers HM, Tomberlin JK, Lambert BD, Kattes D (2008) Development of black soldier fly (Diptera: Stratiomyidae) larvae fed dairy manure. Environ Entomol 37(1):11–15. doi:10.1603/0046-225X(2008)37[11:DOBSFD]2.0.CO;2
Newton GL, Booram CV, Barker RW, Hale OM (1977) Dried Hermetia-illucens larvae meal as supplement for swine. J Anim Sci 44(3):395–400
Nordin A, Nyberg K, Vinnerås B (2009) Inactivation of Ascaris eggs in source-separated urine and feces by ammonia at ambient temperatures. Appl Environ Microbiol 75(3):662–667. doi:10.1128/AEM.01250-08
Paul JW, Beauchamp EG (1989) Relationship between volatile fatty acids, total ammonia, and pH in manure slurries. Biol Wastes 29(4):313–318. doi:10.1016/0269-7483(89)90022-0
R Core Team (2012) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Rockström J, Steffen W, Noone K, Persson Å, Chapin FS, Lambin EF, Lenton TM, Scheffer M, Folke C, Schellnhuber HJ, Nykvist B, de Wit CA, Hughes T, van der Leeuw S, Rodhe H, Sörlin S, Snyder PK, Costanza R, Svedin U, Falkenmark M, Karlberg L, Corell RW, Fabry VJ, Hansen J, Walker B, Liverman D, Richardson K, Crutzen P, Foley JA (2009) A safe operating space for humanity. Nature 461(7263):472–475. doi:10.1038/461472a
Sahlström L, Aspan A, Bagge E, Danielsson-Tham ML, Albihn A (2004) Bacterial pathogen incidences in sludge from Swedish sewage treatment plants. Water Res 38(8):1989–1994. doi:10.1016/j.watres.2004.01.031
Sharpley AN, Chapra SC, Wedepohl R, Sims JT, Daniel TC, Reddy KR (1994) Managing agricultural phosphorus for protection of surface waters: issues and options. J Environ Qual 23(3):437–451. doi:10.2134/jeq1994.00472425002300030006x
Sheppard DC, Newton GL, Thompson SA, Savage S (1994) A value added manure management system using the black soldier fly. Bioresour Technol 50(3):275–279. doi:10.1016/0960-8524(94)90102-3
Sheppard DC, Tomberlin JK, Joyce JA, Kiser BC, Sumner SM (2002) Rearing methods for the black soldier fly (Diptera: Stratiomyidae). J Med Entomol 39(4):695–698. doi:10.1603/0022-2585-39.4.695
Sidhu JPS, Toze SG (2009) Human pathogens and their indicators in biosolids: a literature review. Environ Int 35(1):187–201. doi:10.1016/j.envint.2008.07.006
St-Hilaire S, Sheppard C, Tomberlin J, Irving S, Newton L, McGuire M, Mosley E, Hardy R, Sealey W (2007) Fly prepupae as a feedstuff for rainbow trout, Oncorhynchus mykiss. J World Aquacult Soc 38(1):59–67. doi:10.1111/j.1749-7345.2006.00073.x
Tacon AGJ, Metian M (2008) Global overview on the use of fish meal and fish oil in industrially compounded aquafeeds: trends and future prospects. Aquaculture 285(1–4):146–158. doi:10.1016/j.aquaculture.2008.08.015
UNEP (2010) Waste and climate change: Global trends and strategy framework. United Nations Environmental Program, Osaka
van Huis A, van Itterbeeck J, Klunder H, Mertens E, Halloran A, Muir G, Vantomme P (2013) Edible insects: Future prospects for food and feed security. Food and Agriculture Organization of the United Nations, Rome
Vinnerås B, Björklund A, Jönsson H (2003) Thermal composting of faecal matter as treatment and possible disinfection method—laboratory-scale and pilot-scale studies. Bioresour Technol 88(1):47–54. doi:10.1016/S0960-8524(02)00268-7
Wei J, Jin Y, Sims T, Kniel KE (2009) Survival of human adenovirus 41 in land-applied manure and biosolids. Food Environ Virol 1(3–4):148–154. doi:10.1007/s12560-009-9021-x
Acknowledgments
Funding for this research was provided by the Swedish Research Council (Vetenskapsrådet) as part of their program supporting Swedish research relevant to low- and middle-income countries (U-forsk) and by the Eco-Innovera program SPROUT finance by FORMAS (Sweden) and BAFU (Switzerland). The laboratory work was conducted at the National Veterinary Institute in Uppsala, Sweden.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Lalander, C.H., Fidjeland, J., Diener, S. et al. High waste-to-biomass conversion and efficient Salmonella spp. reduction using black soldier fly for waste recycling. Agron. Sustain. Dev. 35, 261–271 (2015). https://doi.org/10.1007/s13593-014-0235-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13593-014-0235-4