Abstract
Four models based on convolutional neural networks were used to investigate whether image recognition techniques applied to honey bee wings could be used to discriminate among honey bee subspecies. A dataset consisting of 9887 wing images belonging to 7 subspecies and one hybrid was analysed with ResNet 50, MobileNet V2, Inception Net V3, and Inception ResNet V2. Accuracy values of classification of individual wings were over 0.92, and all models outperformed traditional morphometric evaluation. The Inception models achieved the highest accuracies and higher scores of precision and recall for most classes. When wing images were grouped by colony, almost all wings in the colony samples were labelled with the same class. We conclude that automatic image recognition and machine learning applied to honey bee wings can reliably discriminate among the European subspecies and could thus represent a useful tool for fast classification of honey bee subspecies for breeding and conservation aims.
Similar content being viewed by others
1 Introduction
Discrimination of honey bee subspecies is of critical importance for the conservation of honey bee biodiversity. Identification of the A. mellifera subspecies is also important in honey bee breeding because many breeders are keen on keeping pure stocks and often need official certification to attest that their bees belong to a given subspecies.
Knowledge of the geographical distribution of honey bee subspecies is the result of numerous studies based primarily on morphometry. The most complete synthesis on this subject is provided by the monograph of Ruttner (1988), which refers to the application of numerical taxonomy, firstly introduced by DuPraw (1964). Ruttner used a total of 36 characters, including size of various body parts, wing venation, pilosity, and pigmentation, for analysis of honey bee workers from a wide range of geographic regions (Ruttner et al. 1978; Ruttner 1988). This set of characters, known as “standard morphometry,” still constitutes the reference method more often used in studies of geographic variation.
Today, several morphometrical methods of subspecies identification are in use. They differ in precision and time employed for the analysis (Bouga et al. 2011; Meixner et al. 2013). For the purpose of breeding and conservation programmes, rapid and less precise methods, based on the measurement of few characters, are usually preferred, although they are suitable for the discrimination of a limited number of subspecies.
Efforts have been made to facilitate the morphometric identification process towards computerised, and even partially automated, methods. The most suitable body part for computerised analyses is the forewing, where many characters can be rapidly and quite accurately obtained by identification of venation junctions which yield angles and lengths or can be used as landmarks for shape analyses (Bouga et al. 2011; Meixner et al. 2013; Nawrocka et al. 2017; Tofilski 2008). For these kinds of analyses, a scanner, a personal computer, and specific software for data acquisition are required.
A new frontier in the recognition and classification of images is represented by application of artificial intelligence through techniques of machine learning based on neural networks. These systems have recently shown impressive results across a variety of domains and may be particularly suited to solve problems in the field of biological sciences. Computer vision tools have undergone a fast evolution over the past 10 years with the introduction and the subsequent development of Convolutional Neural Networks (hereon CNN) (He et al. 2016). CNNs are extremely flexible in terms of being able to solve many kinds of problems, such as classification, regression, or anomaly detection and in theory are able to learn any computer vision task with high accuracy. Since subspecies recognition can be defined as an image classification problem (Silva et al. 2015), we claim that a CNN could learn to solve it from a set of labelled examples. Thus, the aim of the present study was to investigate whether the modern image recognition techniques could be used to efficiently discriminate among honey bee subspecies through analysis and classification of wing images. These techniques are not new to the field of apidology and have already been employed with success in assessing the status of honey bee comb cells by Alves et al. (2020).
In this paper, we consider four CNN architectures and train them to recognise the honey bee subspecies using a set of 9887 forewing images, belonging to a reference collection containing 7 European subspecies and the Buckfast intraspecific hybrid.
2 Methods
2.1 Wing images, colony samples and morphometrical analyses
The wing images used in this study are stored at CREA-Research Centre for Agriculture and Environment (CREA-AA) and were collected from honey bee colonies over the past 40 years. A total number of 509 honey bee colony samples were used: 273 are part of the reference dataset used at CREA-AA for morphometric subspecies classification, initially validated by means of analysis of several characters according to standard morphometry (Ruttner, 1988); the remaining colony samples were collected by CREA-AA from beekeepers maintaining autochthonous subspecies in Italy and other European countries. These colony samples were chosen based on conformity with the declared subspecies, comparing them to the CREA reference dataset by means of measurement of 30 wing parameters according to the DAWINO (Discriminant Analysis With Numerical Output) method (Meixner et al. 2013), supplemented by check of the third tergite pigmentation, in accordance with the Italian procedure described in Bouga et al. (2011).
The considered honey bee colony samples belong to 7 European subspecies and to the intraspecific hybrid “Buckfast” (from hereon collectively referred to as “classes”) (Table I). Each sample consists of 15–25 right forewings dissected from bees collected from one colony, placed on a photographic slide sheet or a microscope slide and then scanned with a 3200-dpi resolution to obtain digital images.
2.2 Image processing with artificial intelligence
Single-wing pictures (from hereon “images”) were extracted from the above described dataset by means of object detection, performed with a RetinaNet (Lin et al. 2017) model with a ResNet 50 (He et al. 2016) backbone trained on a sample of 22 manually annotated images. Annotation was achieved by creating PASCAL-VOC xml files (Everingham et al. 2010).
Results were manually verified removing false positives (images that were not wings, such as stains on the slide — although this only occurred twice) and obtaining a dataset consisting of eight classes with 9887 images distributed as reported in Table I.
The images were then resized and padded with white pixels to match the square format expected by the considered neural networks described below.
2.3 Recognition models and training
CNN models that are well established in the IT literature and in the industry were considered for the experiment: ResNet 50, MobileNet V2 (Sandler et al. 2018), Inception Net V3 (Szegedy et al. 2016), and Inception ResNet V2 (Szegedy et al. 2017). Though none of these models is to be considered state of the art in the field of image classification (Tan & Le 2019), they all provide reasonably good overall performance and they have widely available implementations in a number of deep learning packages such as TensorFlow, Keras, and PyTorch.
Each one of the considered models presents a substantial complexity, with millions of parameters to be learned during training (Mobile Net V2 has roughly 3.4 million trainable parameters, and it is the smallest model considered herein); hence, it is of vital importance to optimise the training procedure to achieve good results in acceptable times. A stratified cross-validation (Zeng & Martinez, 2000) procedure was used, with 10 folds (Figure 1). Such a procedure is iterative in its nature and well established in the Machine Learning community. The data was randomly partitioned in ten equal sized subsets (folds), each one respecting the original dataset’s class proportions: i.e., the most represented class remained the most represented class across all the ten folds, and the other classes represented proportionally. Once folds were computed, an iterative process began and at each iteration the partitions built in the previous step were assembled into three larger partitions: training set, validation, set and test set, which are referred to as splits. The training set was made of 8 folds of the data, and the other two splits of a single fold each. Each one of the splits, being either a fold or the union of eight folds, respected the class proportions of the original dataset. At each iteration, a new model instance was trained over the training set and evaluated on the test set, producing a set of class predictions for each image in the latter split. The procedure was repeated until each fold was used as test set once, which means that each image in the dataset received a model prediction by a model that was not fed with it during its training. We used such predictions to evaluate model performance metrics over the whole dataset.
The above-described experimental design was extended with data bootstrapping and early stopping during training to address two criticalities: overfitting and statistical bias. The imbalance in the distribution of classes presents a significant statistical bias towards more represented subspecies, such as Siciliana; to address this issue, the data included in the training split was bootstrapped to obtain a more regular distribution, thus reducing statistical bias during model training. All classes in the training split were randomly resampled with replacement until each of them consisted in at least 1600 images, that corresponds to 0.6 times the cardinality of the most numerous class. Test and validation data were not bootstrapped to avoid tampering model selection and evaluation. This process is summarised in (Figure 2), where, for the sake of readability, four folds instead of ten are pictured.
Of the three splits, the bootstrapped training set was used to feed the model during training, the validation set was used to check model progress during training, and finally, the test set was used to perform model evaluation. The training procedure we used for all models was stochastic gradient descent (Bottou 2010) (hereon SGD) with triangular learning rate scheduling (Smith 2017). This is an iterative procedure also, and it requires training data to be processed multiple times, each one called epoch. Due to its iterative nature, the training procedure could, theoretically, go on forever and it is up to the data scientist to stop it when an appropriate fit is reached. Since it is impossible to know a priori the optimal number of training epochs, we determined it empirically by introducing the validation set. The training procedure was stopped when the performance metric applied to the validation set (accuracy, see the “Analysis” section) reached a maximum, and no further progress could be observed. Such a maximum point can be considered as the best fit, since it reasonably provides a sweet spot between underfitting and overfitting. Since the validation set was used to tune the number of training epochs, its data was, as a matter of fact, embedded in the trained model, even though not actually processed at training time; hence, the need for a third split to perform an un-biased evaluation.
All models were trained for twenty epochs with online data augmentation, which means generating multiple versions of the same image as it is passed to the model during training. This solution, with respect to a pre-computed set of perturbated images, had two advantages: it was more memory efficient (less images to be loaded in the GPU memory) and introduced a higher degree of randomisation over different epochs, allowing the model to achieve a higher tolerance towards sub-optimal images. Training images were augmented by means of random rotations, vertical flip, horizontal flip, and brightness adjustment to increase variability in the training data, with replication padding to avoid disrupting the original pixel colour distributions. The various transformations were applied stochastically in cascade, meaning that a wing image could be, for instance, both flipped and rotated, to maximise the randomness of transformations and, hopefully, the model robustness against noisy data.
2.4 Analysis
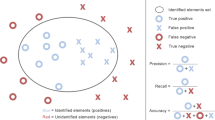
Three well-known classification metrics (Powers 2011) were used to assess model performance:
-
Accuracy: the fraction of correctly classified images. It is used to evaluate overall model performance.
-
Precision: also known as positive predictive value, it is the fraction of positive values that are true positives. It is used to evaluate model performance with respect to a given class. It represents a measure of how good the model is at avoiding false positives (e.g. considering A. m. ligustica as declared class, a false positive is a wing classified as A. m. ligustica which actually is A. m. carnica)
-
Recall: also known as specificity, it is the fraction of positive samples correctly identified by the system. It is used to evaluate model performance with respect to a given class. It represents a measure of how good the model is at avoiding false negatives (e.g. considering A. m. ligustica as declared class, a false negative is a wing classified as A. m. carnica which actually is A. m. ligustica).
Precision and recall, being complementary with each other, are frequently accompanied by their harmonic mean, called F1 score, which is useful when a balance between precision and recall is required.
3 Results
3.1 Model evaluation
The accuracy results for all the four considered models are shown in Table II. Values were calculated for each test split considered in the cross-validation procedure and then averaged. The Inception ResNet and Inception Net architectures appeared to perform better than the ResNet and MobileNet ones, with higher average accuracy and lower accuracy variance.
Due to our experimental design, our cross-validation procedure was built on pre-computed stratified partitions of the data set; hence, we can state that each image was present in the test partition of the data exactly once, allowing us to make a union of the test splits predictions and evaluate global metrics over the 10 replicas of all models considered. By performing this aggregation, we lost some distributional information; however, as shown in Table II, the accuracy variance among different folds was < 0.01 for Inception Net V3 and Inception ResNet V2 and < 0.02 for MobileNet and < 0.04 for ResNet, implying that all models achieved rather homogenous performance over different folds, and more importantly, since classes were not evenly represented in our data, it allowed us to evaluate metrics on a relevant-sized sample for all the considered classes.
To better understand the performance difference between the former two high-accuracy models, we evaluated, for each considered class, the precision, recall, and F1 values over the union of test splits (Table III). The Inception ResNet model appeared to achieve slightly higher scores in most classes; however, the performance of these two models appeared to be consistent across classes. Most scores were over 0.99, and with both models, the classes Caucasica, Iberiensis, and Anatoliaca were classified with the highest precision and recall.
To further analyse classification errors, we arranged the predictions made by the tested models into confusion matrices, i.e. matrices where rows represent ground truth values and columns model predictions. As suggested by the precision and recall metrics, the confusion matrices (Figure 3) were sparse and almost diagonal, with few non-zero elements outside the diagonals. Looking at the non-diagonal cells, we can observe how the two models were consistent in the errors, both most frequently mistaking Siciliana and Carnica for Ligustica. In addition, the Inception Net V3 model was more likely to mistake Ligustica for Buckfast or Carnica, while the Inception ResNet V2 model appeared to be less prone to this type of error.
The analysis presented so far are relative to the performance of models on a single-wing image basis; however, bee wings are usually contained in samples that contain multiple wings (in our data set usually 18) coming from different individuals of a same colony. To assess how the computer vision models would perform in a real world scenario, in post-processing we grouped the single images by colony, and considered the mode class of the individual wing predictions for assignment of the colony to one of the eight classes. This process resulted in a colony-wise accuracy ranging from 0.9921 to 1, as shown in Table IV. These values suggest that the error observed when classifying a single wing image is distributed over different colony samples and that all the four considered models could correctly classify the majority of the wings belonging to a same colony. To further illustrate this fact, we evaluated for each colony a confidence value defined as the fraction of wings of the colony sample belonging to the mode class. In Figure 4, we show, for each tested model, the distribution of these confidence values among our considered colony samples. For all four models, the most common scenario was the one wherein all wings were labelled with the same class; however, a clear difference between the top scoring networks (Inception ResNet V2 and Inception Net V3) and the lower scoring ones could be noticed: the top scoring networks had a much shorter tail of low confidence samples, while the lower scoring networks had longer tails going down to 0.4 confidence and below, and included a non-negligible number of colony samples.
3.2 Comparison with morphometric method
To exactly compare the morphometric approach used at CREA and the computer vision models, a subset of 242 colony samples was extracted from the considered dataset and all individual wings were labelled with the morphometric approach based on measurement of 30 characters according to DAWINO method (Meixner et al, 2013), with respect to the considered eight classes reported in Table I. At colony level, i.e. by considering the mode class of bee wings for each colony sample (as described above), both the morphometric procedure and the computer vision models produced accurate predictions, with the morphometric approach achieving a 0.979 accuracy, with only five misclassified colonies, and all four Computer Vision models, a 1.0 accuracy, meaning that all the 242 colony samples were correctly labelled. The confusion matrixes produced by the morphometric method and Inception ResNet V2 at the colony level are shown in (Figure 5a, b).
Confusion matrices relative to a subset of 242 colony samples which were classified with the morphometric approach (baseline) and with the best preforming computer vision model (Inception ResNet V2), with respect to the considered eight classes. a Morphometric classification at colony level. b Inception ResNet V2 classification at colony level. c morphometric classification at individual wing level. d Inception ResNet V2 classification at individual wing level
To gain additional insights on the compared performance, the individual wing labelling task was also considered. The bee wings in the colony samples were classified individually with the morphometric method, resulting in the confusion matrix shown in (Figure 5c), and an accuracy of 0.8607 (Table V). On the same set of 242 colony samples, the computer vision approaches were evaluated and produced accuracy results ranging from 0.9289 (MobileNet) to 0.9915 (Inception Net) (Table V). The confusion matrix produced by the Inception Net V3 model is shown in (Figure 5d). Looking at the accuracy values scored by the various models in our benchmark, we can see that Inception Net and Inception ResNet show virtually identical performances, while ResNet and MobileNet appeared to be substantially inferior in terms of accuracy. The considerations on accuracy can be extended to class-wise metrics as well, with computer vision models achieving substantially higher scores for every class, as shown in Table VI.
Another notable aspect is that the computer vision pipeline considered significantly more images for almost all classes (Table VII).
4 Discussion
Beekeepers and breeders are ever more conscious of the importance of the genetic origin of their honey bees, due to a more widespread environmentally sensitive awareness, and partly due to the phenomenon of colony losses which hit honey bees in the last decade (Brodschneider et al., 2016; Neumann & Carreck, 2010; Ratnieks & Carreck, 2010; Zee et al., 2014). The main culprits of these losses have been universally acknowledged as a combination of factors, including pesticides (Goulson et al., 2015), parasites (Di Prisco et al., 2016), poor nutrition (Brodschneider & Crailsheim, 2010; Di Pasquale et al., 2016), and genetic origin (Büchler et al., 2014). For these reasons, many beekeepers across Europe are interested in checking whether the bees they are managing correspond to the autochthonous subspecies. In some cases, a certification of the subspecies will enable them to receive subsidies according to local or European legislation; in other cases, it may enable access to conservation areas, or it may increase the market value of their stock. A fast and cheap method would, therefore, be a useful tool for the apicultural sector.
With the results of this study, we confirmed our hypothesis that image recognition by Convolutional Neural Networks applied to honey bee wings can be used to discriminate between subspecies, even more successfully than the morphometrical analysis. Notably, by analysing 9887 wing images, representing seven Apis mellifera subspecies and one intraspecific hybrid, we obtained discrimination with an accuracy ranging between 0.92 and 0.99, with the four considered network architectures: ResNet50, MobileNet V2, Inception Net V3 and Inception ResNet V2. The latter model was the one with the best performance, achieving scores of precision, recall, and F1 higher than 0.98 for all classes.
It is interesting to notice that the misclassifications were mostly within evolutionary lineage and between geographically close subspecies. Similar misclassifications were found in a recent study using genetic markers (Momeni et al., 2021).
When the morphometric method was applied to classify individual wings which were also classified by the CNN models, we found that its accuracy was notably lower, confirming the discriminatory power of the computer vision method. In a previous study involving 5 European subspecies and populations, the classical morphometric method performed with a maximum accuracy of 0.94 and as low 0.33 for some classes, including Ligustica, although it must be noted that the samples had not been chosen based on conformity to race standards as in our case (Francis et al., 2014). It is also worth noticing how the number of considered single wing images varied between the morphometric method and the computer vision one, with differences in class numerosity ranging from 7 to 60%. This is an effect of the two different processing pipelines involved: in the morphometric approach, images were manually inspected by a human expert that handpicked a set of representative wings and proceeded to classify them, while in the computer vision pipeline, the images were processed by the RetinaNet model that passed to the classification model whatever it identified as a bee wing, without excluding blurred, noisy, or damaged images.
In characterisation studies following publication of Ruttner (1988) (Meixner, et al., 2011, 1994; Miguel et al., 2011; Sheppard et al., 1997; Sheppard & Meixner, 2003), standard morphometry parameters were considered as the chief method or the baseline which accompanies other methods, because the statistical method using morphometrical measures was the one used for classification of the genus Apis at the specific and subspecific level. Also, as shown by (Henriques et al., 2020), morphometry can identify even complex genetic patterns, offering a reliable and low-cost alternative for preliminary estimation of population structure. The method we developed here can effectively differentiate several subspecies as described by Ruttner (1988), and is validated with a unique dataset of reference samples stored at CREA-AA and collected in the last 40 years. Moreover, with regard to methods based on wing venation measurement proposed so far, although software programs have been developed to simplify the procedure for classical and geometric morphometry, the operator still plays a certain role in identifying and checking vein intersections. On the contrary, the fast method proposed here excludes any operator intervention in the analytical process and the wing recognition and classification relies totally on a system of computer vision joined with an artificial intelligence software, avoiding any processing of a predefined geometric pattern.
Molecular techniques are developing fast but are not yet easily available for beekeepers. Furthermore, methods based on honey bee maternal lines through the mtDNA sequencing, resulting in the production of different patterns of mitotypes, although interesting in terms of phylogenetic studies (Ilyasov et al., 2019) are not exhaustive in terms of subspecies recognition (Meixner et al., 2013). Only the newest techniques of gDNA sequencing and SNPs analysis (Momeni et al., 2021; Spötter et al., 2012; Whitfield et al., 2006) have shown to be able to discriminate almost all subspecies. Recently, a molecular tool based on SNPs having as a reference almost 4000 individual honey bees, originating from samples where the subspecies assignment was also confirmed with the morphometric analysis, has been developed, and is commercially available (Momeni et al., 2021). Although this tool marks a breakthrough, the cost per representative colony sample (10–20 bees) is still expensive for the average beekeeper. For this reason, our innovative approach based on wing image analysis could represent a fast and cheap alternative to molecular tools, and could even be seen as a complementary method, in which many individuals per colony are considered, while the molecular tool could be based on a single individual per colony.
Image recognition technology is advancing at a fast pace, and machine learning has enabled continuous training of models on high numbers of samples. Production-grade models such as the ones we tested in this study appear to be powerful enough to achieve a satisfactory accuracy and are available in several widespread software packages available on the market. In conclusion, the analyses performed in this study show that automatic image recognition and machine learning, applied to honey bee wings, can discriminate subspecies. More specifically, the most promising CNN model applied to wing samples was Inception Resnet V2.
Our results underscore the role that image recognition applied to the biological systems can play. We hereby show that image recognition can represent a cutting edge and useful tool to provide breeders and beekeepers with a fast and cheap classification of their honey bees, setting the basis for future development of a hands-on application for conventional informatic devices.
Availability of data and material
The wing images are available from the corresponding author on reasonable request.
References
Alves, T. S., Pinto, M. A., Ventura, P., Neves, C. J., Biron, D. G., Junior, A. C., De Paula Filho, P. L., Rodrigues, P. J. (2020) Automatic detection and classification of honey bee comb cells using deep learning. Comput. Electron. Agric. 170, 105244. https://doi.org/10.1016/j.compag.2020.105244
Bottou, L. (2010) Large-Scale Machine Learning with Stochastic Gradient Descent. Proc. COMPSTAT’2010. 177–186. https://doi.org/10.1007/978-3-7908-2604-3_16
Bouga, M., Alaux, C., Bienkowska, M., Büchler, R., Carreck, N. L., et al. (2011) A review of methods for discrimination of honey bee populations as applied to European beekeeping. J. Apic. Res. 50(1), 51–84. https://doi.org/10.3896/IBRA.1.50.1.06
Brodschneider, R., & Crailsheim, K. (2010). Nutrition and health in honey bees. Apidologie, 41(3), 278–294. https://doi.org/10.1051/apido/2010012
Brodschneider, R., Gray, A., van der Zee, R., Adjlane, N., Brusbardis, V., Charrière, J.-D., … Woehl, S. (2016). Preliminary analysis of loss rates of honey bee colonies during winter 2015/16 from the COLOSS survey. Journal of Apicultural Research, 55(5), 375–378. https://doi.org/10.1080/00218839.2016.1260240
Büchler, R., Costa, C., Hatjina, F., Andonov, S., Meixner, M. D., Le Conte, Y., … Wilde, J. (2014). The influence of genetic origin and its interaction with environmental effects on the survival of Apis Mellifera L. Colonies in Europe. Journal of Apicultural Research, 53(2), 205–214. https://doi.org/10.3896/IBRA.1.53.2.03
Di Pasquale, G., Alaux, C., Le Conte, Y., Odoux, J.-F., Pioz, M., Vaissière, B. E., … Decourtye, A. (2016). Variations in the Availability of Pollen Resources Affect Honey Bee Health. PLOS ONE, 11(9), e0162818. https://doi.org/10.1371/journal.pone.0162818
Di Prisco, G., Annoscia, D., Margiotta, M., Ferrara, R., Varricchio, P., Zanni, V., … Pennacchio, F. (2016). A mutualistic symbiosis between a parasitic mite and a pathogenic virus undermines honey bee immunity and health. Proceedings of the National Academy of Sciences, 113(12), 3203–3208. https://doi.org/10.1073/pnas.1523515113
DuPraw, E. J. (1964) Non-Linnean Taxonomy. Nature. 202(4935), 849–852. https://doi.org/10.1038/202849a0
Everingham, M., Van Gool, L., Williams, C. K. I., Winn, J., & Zisserman, A. (2010). The pascal visual object classes (VOC) challenge. International Journal of Computer Vision, 88(2), 303–338. https://doi.org/10.1007/s11263-009-0275-4
Francis, R. M., Kryger, P., Meixner, M., Bouga, M., Ivanova, E., Andonov, S., … Wilde, J. (2014). The genetic origin of honey bee colonies used in the coloss genotype-environment interactions experiment: A comparison of methods. Journal of Apicultural Research, 53(2). https://doi.org/10.3896/IBRA.1.53.2.02
Goulson, D., Nicholls, E., Botias, C., & Rotheray, E. L. (2015). Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science, 347(6229), 1255957–1255957. https://doi.org/10.1126/science.1255957
He, K., Zhang, X., Ren, S., Sun, J. (2016) Deep Residual Learning for Image Recognition. 2016 IEEE Conf. Comput. Vis. Pattern Recognit. 770–778. https://doi.org/10.1109/CVPR.2016.90
Henriques, D., Chávez-Galarza, J., S. G. Teixeira, J., Ferreira, H., J. Neves, C., Francoy, T. M., & Pinto, M. A. (2020). Wing Geometric Morphometrics of Workers and Drones and Single Nucleotide Polymorphisms Provide Similar Genetic Structure in the Iberian Honey Bee (Apis mellifera iberiensis). Insects, 11(2), 89. https://doi.org/10.3390/insects11020089
Ilyasov, R. A., Nikolenko, A., Tuktarov, V., Goto, K., Takahashi, J.-I., & Kwon, H. W. (2019). Comparative analysis of mitochondrial genomes of the honey bee subspecies A. m. caucasica and A. m. carpathica and refinement of their evolutionary lineages. Journal of Apicultural Research, 58(4), 567–579. https://doi.org/10.1080/00218839.2019.1622320
Lin, T., Girshick, R., Doll, P., Goyal, P., Girshick, R., He, K., Dollar, P. (2017) Focal Loss for Dense Object Detection ar. Proc. IEEE Int. Conf. Comput. vision. 2980–2988. https://doi.org/10.1109/ICCV.2017.324
Meixner, M. D., Leta, M. A., Koeniger, N., & Fuchs, S. (2011). The honey bees of Ethiopia represent a new subspecies of Apis mellifera—Apis mellifera simensis n. ssp. Apidologie, 42(3), 425–437. https://doi.org/10.1007/s13592-011-0007-y
Meixner, M. D., Pinto, M. A., Bouga, M., Kryger, P., Ivanova, E., & Fuchs, S. (2013). Standard methods for characterising subspecies and ecotypes of Apis mellifera. Journal of Apicultural Research, 52(4), 1–28. https://doi.org/10.3896/IBRA.1.52.4.05
Meixner, M. D., Sheppard, W. S., Dietz, A., & Krell, R. (1994). Morphological and allozyme variability in honey bees from Kenya. Apidologie, 25(2), 188–202. https://doi.org/10.1051/apido:19940207
Miguel, I., Baylac, M., Iriondo, M., Manzano, C., Garnery, L., & Estonba, A. (2011). Both geometric morphometric and microsatellite data consistently support the differentiation of the Apis mellifera M evolutionary branch. Apidologie, 42(2), 150–161. https://doi.org/10.1051/apido/2010048
Momeni, J., Parejo, M., Nielsen, R. O., Langa, J., Montes, I., Papoutsis, L., … Estonba, A. (2021). Authoritative subspecies diagnosis tool for European honey bees based on ancestry informative SNPs. BMC Genomics, 22, 101 (2021). https://doi.org/10.1186/s12864-021-07379-7.
Nawrocka, A., Kandemir, İ., Fuchs, S., Tofilski, A. (2017) Computer software for identification of honey bee subspecies and evolutionary lineages. Apidologie. 49, 172–184. https://doi.org/10.1007/s13592-017-0538-y
Neumann, P., & Carreck, N. L. (2010). Honey bee colony losses. Journal of Apicultural Research, 49(1), 1–6. https://doi.org/10.3896/IBRA.1.49.1.01
Powers, D. M. W. (2011) Evaluation: from precision, recall and f-measure to roc, informedness, markedness & correlation. J. Mach. Learn. Technol. 2(1), 37–63. http://bioinfopublication.org/viewhtml.php?artid=BIA0001114
Ratnieks, F. L. W., & Carreck, N. L. (2010). Clarity on honey bee collapse? Science, 327(5962), 152–153. https://doi.org/10.1126/science.1185563
Ruttner, F., Tassencourt, L., Louveaux, J. (1978) Biometrical-statistical analysis of the geographic variability of apis mellifera L. I. material and methods. Apidologie. 9(4), 363–381. https://doi.org/10.1051/apido:19780408
Ruttner, Friedrich. (1988) Biogeography and Taxonomy of Honeybees. Springer-Verlag, Germany
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., Chen, L. C. (2018) MobileNetV2: Inverted Residuals and Linear Bottlenecks. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. https://doi.org/10.1109/CVPR.2018.00474
Sheppard, W. S., Arias, M. C., Grech, A., & Meixner, M. D. (1997). Apis mellifera ruttneri, a new honey bee subspecies from Malta. Apidologie, 28(5), 287–293. https://doi.org/10.1051/apido:19970505
Sheppard, W. S., & Meixner, M. D. (2003). Apis mellifera pomonella , a new honey bee subspecies from Central Asia. Apidologie, 34(4), 367–375. https://doi.org/10.1051/apido:2003037
Silva, F. L. da., Grassi Sella, M. L., Francoy, T. M., Costa, A. H. R. (2015) Evaluating classification and feature selection techniques for honeybee subspecies identification using wing images. Comput. Electron. Agric. 114, 68–77. https://doi.org/10.1016/j.compag.2015.03.012
Smith, L. N. (2017) Cyclical learning rates for training neural networks. Proc. - 2017 IEEE Winter Conf. Appl. Comput. Vision, WACV 2017. https://doi.org/10.1109/WACV.2017.58
Spötter, A., Gupta, P., Nürnberg, G., Reinsch, N., & Bienefeld, K. (2012). Development of a 44K SNP assay focussing on the analysis of a varroa-specific defence behaviour in honey bees (Apis mellifera carnica). Molecular Ecology Resources, 12(2), 323–332. https://doi.org/10.1111/j.1755-0998.2011.03106.x
Szegedy, C., Ioffe, S., Vanhoucke, V., Alemi, A. (2017) Inception-v4, inception-ResNet and the impact of residual connections on learning. AAAI’17 Proc. Thirty-First AAAI Conf. Artif. Intell.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z. (2016) Rethinking the Inception Architecture for Computer Vision. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. https://doi.org/10.1109/CVPR.2016.308
Tan, M., Le, Q. V. (2019) EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. 36th Int. Conf. Mach. Learn. ICML 2019. 10691–10700. http://arxiv.org/abs/1905.11946
Tofilski, A. (2008) Using geometric morphometrics and standard morphometry to discriminate three honeybee subspecies. Apidologie. 39(5), 558–563. https://doi.org/10.1051/apido:2008037
Whitfield, C. W., Behura, S. K., Berlocher, S. H., Clark, A. G., Johnston, J. S., Sheppard, W. S., … Tsutsui, N. D. (2006). Thrice out of Africa: Ancient and recent expansions of the honey bee, Apis mellifera. Science, 314(5799), 642–645. https://doi.org/10.1126/science.1132772
Zee, R. van der, Brodschneider, R., Brusbardis, V., Charrière, J.-D., Chlebo, R., Coffey, M. F., … Gray, A. (2014). Results of international standardised beekeeper surveys of colony losses for winter 2012–2013: analysis of winter loss rates and mixed effects modelling of risk factors for winter loss. Journal of Apicultural Research, 53(1), 19–34. https://doi.org/10.3896/IBRA.1.53.1.02
Zeng, X., & Martinez, T. R. (2000). Distribution-balanced stratified cross-validation for accuracy estimation. Journal of Experimental and Theoretical Artificial Intelligence, 12(1), 1–12. https://doi.org/10.1080/095281300146272
Acknowledgements
We want to thank Dr Rui Gonçalves (Department of Zoology at University of Oxford, UK) and Dr Tiago Moreira (APICAVE — Associação de Apicultores do Cavado e do Ave, Maximinos, Braga, Portugal); Dr. Padruot M. Fried and Verein Schweizerischer Mellifera Bienenfreunde (http://www.mellifera.ch) (Switzerland) and Dr Alessandro Manzano (Associazione per l’Ape Carnica Friulana, Italy) for their help in providing the additional A. m. iberiensis, A. m. mellifera, and A. m. carnica samples used in the study. We also appreciate the support given by Dr. Dariusz Gerula (Research Institute of Horticulture Apicultural Division, Pulawy, Poland) and Prof. Aleksandar Uzunov (Ss. Cyril and Methodius University in Skopje, North Macedonia) in providing extra wing images used for internal validation of the system.
Funding
The present study was performed under the programs “Tenuta dei libri genealogici” and AGRIDIGIT funded by Italian Ministry of Agriculture, Dipartimento delle politiche europee ed internazionali e dello sviluppo rurale.
Author information
Authors and Affiliations
Contributions
D.D.N. conceived this research and analysed the data with the CNN models; E.C. analysed the data with the morphometrical approach. C.C., G.D.P., and E.C. collected and validated the samples; all authors contributed to writing the paper.
Corresponding author
Ethics declarations
Ethics
Ethical approval was not required for this study as the experimental work was conducted with unregulated invertebrate species.
Consent to participate and publication
Beekeepers who contributed samples consented for the outcoming data to be published in this study.
Conflict of interest
The authors declare no competing interests.
Additional information
Manuscript editor: Marina Meixner
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
De Nart, D., Costa, C., Di Prisco, G. et al. Image recognition using convolutional neural networks for classification of honey bee subspecies. Apidologie 53, 5 (2022). https://doi.org/10.1007/s13592-022-00918-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13592-022-00918-5