Abstract
Austroplebeia Moure is an Australian stingless bee genus. The current descriptions for the species within this genus are inadequate for the identification of specimens in either the field or the laboratory. Here, using multiple diagnostic methodologies, we attempted to better delimit morphologically identified groups within Austroplebeia. First, morphological data, based on worker bee colour, size and pilosity, were analysed. Then, males collected from nests representing morphologically similar groups were dissected, and their genitalia were imaged using light microscopy and scanning electron microscopy. Next, data for the geometric morphometric analysis of worker wing venations were obtained. Finally, molecular analysis, using mitochondrial 16S ribosomal RNA (16S rDNA), was conducted on workers from representative nests for each group which displayed morphological similarities. Data deriving from the four different diagnostic approaches were compared, resulting in the separation of two groups, plus an unresolved species complex.
Similar content being viewed by others
1 Introduction
Worldwide, there is a dearth of taxonomic information on insect pollinators (Dias et al. 1999; Imperatriz-Fonseca 2010). Many Australian bee species are undescribed, with over half of the named taxa requiring revision (Batley and Hogendoorn 2009). The Australian stingless bee species within the genus Austroplebeia Moure are amongst this group.
At present, the genus Austroplebeia has nine named species (Cardale 1993). The taxonomic classifications were based on a small number of pinned specimens, many of which were collected from flowers a century or more ago (Cockerell 1905; Cockerell 1910). These species, together with the primary type collection locations and an abbreviated morphological description, can be found in Appendix 1 of the supplementary online material.
The published descriptions of species within Austroplebeia are inadequate and insufficient for accurate specimen identification. Different sets of characters are described for each species. For instance, Mocsáry (Friese 1898) stated that Austroplebeia cincta and Austroplebeia australis differ in the shininess of the integument; Cockerell (1905) stated that Austroplebeia essingtoni and A. australis differ in the colouration of the scutellum, apical terga and clypeus, and Rayment (1932) stated that Austroplebeia websteri and Austroplebeia cassiae differ in the length of the flagellum and the colour of the legs. Rasmussen (2008) stated that “no workable key exists for Austroplebeia.”
The species within Austroplebeia require redescription; however, this was a task outside the scope of the current study. Our broad aims were to place specimens of Austroplebeia workers into groups, delimit these groups into species and, ultimately, provide a framework to redescribe the species within the genus Austroplebeia. A number of reliable diagnostic techniques were combined, including morphological characteristics such as size and colour, wing vein morphometrics, male genital morphology and molecular analysis.
2 Materials and methods
2.1 Specimen collection
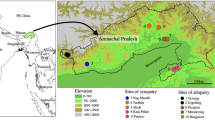
To achieve an accurate description of species, it is important to adequately sample populations, incorporating a large geographic range (Vogler and Monaghan 2007). Between 1980 and 1998, nine field trips were conducted by Les and Anne Dollin throughout Australia, covering Western Australia (WA), the Northern Territory (NT), Queensland (QLD) and New South Wales (NSW). Areas visited were chosen according to where primary type specimens had been collected (Figure 1). Workers were collected from over 106 Austroplebeia nests. To ensure that males earmarked for identification were collected in association with worker nestmates, males were collected from opened nests. Bees were killed in ethyl acetate vapour killing jars and either pinned on site or dry-stored in specimen jars with tissue paper.
Locations of holotype specimens collected between ca. 1840 and 1931 (listed in Appendix 1).
Seven pinned specimens of New Guinean (NG) (two from Papua, Indonesia, and five from Papua New Guinea) A. cincta workers were identified by and obtained from the late Professor Shôichi F. Sakagami (Hokkaido University, Sapporo, Japan) and added to the Austroplebeia collection. At the time of field sampling, it was thought that A. cincta only occurred in NG. In 2009, a bee which had similar markings to A. cincta was found in the Daintree area of northern QLD. Subsequently, a series of worker specimens was collected from six additional nests. The distinctive markings and hair patterns on the faces and thoraces matched those of the NG A. cincta. The specimens were tentatively identified as A. cincta; however, they are referred to as the ‘cincta’ group in this paper.
The Austroplebeia samples analysed in this study were divided into six groups on the basis of morphological features that are further explained in the results: ‘symei’, ‘intermediate’, ‘australis’, ‘striped’, ‘curved’ and ‘cincta’.
2.2 Colouration analysis and head width of indigenous Austroplebeia groups
A sample (n = 10–20) of workers was available from 71 of the 106 sampled nests. These specimens were used in the analysis of body colouration and head width (HW) measurement.
Thoracic markings were digitally imaged using a Leica MZIZ stereomicroscope (Leica Microsystems, North Ryde, Australia) at ×20 magnification. Photographic records were obtained with a JVC digital camera (KY F1030, Victor Company of Japan, Ltd). Using Image-Pro Plus 5.1 software (MediaCybernetics, Bethesda, MD, USA), the total area of the colour markings and the total area of the thorax of each bee were outlined and measured. The proportion of the thorax with colour markings for each ‘typical’ colour grade was calculated as follows: The proportion of marking (%) = (Total area of colour / Total area of thorax) × 100.
The colour grade of 10–20 bees in each nest (n = 71) was recorded, and a mean colour score was calculated for each nest.
For each nest (n = 71), the mean HW was plotted against the mean colour score. The analyses did not include the ‘cincta’ group, as they were not available at the time. Characteristics were analysed by one-way ANOVA, and means were compared by Tukey’s HSD test in SPSS 17 (IBM Corporation, Armonk, NY, USA). The setting of significance was α = 0.05. Data for both HW and colour score were tested prior to analysis for normality using Shapiro-Wilk’s test and for homogeneity of variance using Levene’s test.
2.3 Austroplebeia cincta morphology
The seven, previously identified A. cincta specimens from NG and the new ‘cincta’ specimens from the Daintree were imaged and photographed at ×20 magnification, using stereomicroscopy. All of the Austroplebeia groups were compared for differences in morphological characteristics.
2.4 Male morphology
No A. cincta or ‘cincta’ specimens were available for the male morphology studies, but males were available for all indigenous groups.
Dissection and identification of taxonomic characters in the male genital structures can be reliable species delimiting methods (Michener 2000). Dollin et al. (1997) demonstrated taxonomic differences in Australian Trigona (now Tetragonula) spp. through dissection and comparison of terminal segments of male specimens.
The genitalia of two to four males from each group (n = 19) were cleared in 1 % potassium hydroxide for 48 h and then dissected under a Wild M5 dissecting microscope, at ×32 magnification.
Specimens were imaged with a JSM-6510LV scanning electron microscope (SEM; JEOL, Japan). A high resolution of 3.0 nm was achieved at high vacuum mode, 30 kV, GUI interface. Detailed preparation techniques for male genitalia are provided in Appendix 2 in the supplementary online material. Differences detected in these images prompted closer examination of protruding genital structures in stored specimens using stereomicroscopy.
2.5 Geometric morphometry of wing venations
As a result of preliminary studies, the ‘intermediate’ group was not included in this analysis. Specimens were prepared for geometric morphometric analysis, according to the methodology proposed by Francoy et al. (2011). Ten homologous landmarks were plotted at the wing vein intersections, within the central part of the wing (Figure 2) using tpsDig2 V2.16 software (Rohlf 2005). The rest of the analysis was conducted according to the protocol proposed in Francoy et al. (2008). The statistical analysis was carried out using Statistica 6.0 (StatSoft 2001). The Mahalanobis distances between the centroids of the groups were also calculated in the discriminant analysis, and a dendogram of morphological similarity was constructed based on these data. The dendogram was constructed using neighbour-joining methodology (Saitou and Nei 1987) and MEGA version 4.1 (Tamura et al. 2007). The coefficients of cophenetic correlation, using TreeFit (Kalinowsi 2009), were calculated to determine if the dendogram gave a good representation of these data. Detailed preparation and analysis techniques for the wing morphometric studies can be found in Appendix 3 in the supplementary online material.
By the time this study was underway, ‘cincta’ nests from the Daintree had been sampled and wings from this group, as well as the NG A. cincta, were included.
2.6 DNA extraction, amplification and sequencing
Extraction of DNA from the legs and thoracic tissue of 53 specimens, most of which were dried and up to 27 years old, was carried out using QIAamp DNA Micro Kits (Qiagen, Hilden, Germany). The manufacturer’s protocol was followed, with some modifications: Tissue was lysed overnight at 56 °C and 300 rpm on an Eppendorf Thermomixer comfort (Eppendorf South Pacific, North Ryde, Australia) and elution was with 60 μL of elution buffer.
Several DNA amplification protocols were used in an attempt to obtain the best templates. Cytochrome oxidase I (COI) primers successfully amplified DNA from fresh specimens of ‘australis’ and ‘symei’; however, amplification of the old, dried ‘symei’ specimens was unsuccessful. Data from COI were unavailable for inclusion in this study.
Primers for mitochondrial 16S ribosomal RNA (16S rDNA), LR13943F (5′-CACCTGTTTATCAAAAACAT-3′) and LR13392R (5′-CGTCGATTTGAACTCAAATC-3′), using the protocol described by Costa et al. (2003) successfully amplified a 420-bp amplicon for both fresh and old specimens of ‘australis’ and ‘symei’ and were therefore utilised for the remaining studies. Detailed preparation techniques for DNA template amplification can be found in Appendix 4 in the supplementary online material.
Sequences were assembled, analysed and manually adjusted using Sequencher® 4.5 (Gene Codes Corporation, Ann Arbor, MI, USA). Initially, sequence similarities with previously published data were obtained through an online BLAST® search (Altschul et al. 1990). This confirmed that the sequences were from mitochondrial 16S rDNA of Austroplebeia. Out of a total of 96 amplification attempts from 53 specimens, only 31 readable sequences were obtained.
2.7 Phylogenetic analysis
The data analysis and resulting phylogenetic tree construction were performed, using 31 sequences (accession numbers can be found in Appendix 5 in the supplementary online material). Each nest had previously been allocated to one of six morphological groups, as part of the HW and colour analysis. Each sequence was identified by its nest code, consisting of the morphological group name and the original collection code. 16S rDNA sequences for each specimen were aligned for comparison using MUSCLE within MEGA 5.2 (Tamura et al. 2011). Additional Austroplebeia 16S rDNA sequences were obtained from the GenBank database (http://www.ncbi.nlm.nih.gov/nuccore) and included in the phylogenetic analysis. Alignments were compared using overall mean distance (p-distance model; MEGA 5.2) to determine whether the alignments generated were sufficiently reliable to produce a phylogenetic tree. The criterion of at least 66 % sequence identity was used for DNA sequences to ensure a minimum of 50 % alignment accuracy (Kumar and Filipski 2007). Only 315 bp were included in the alignment, due to trimming for a number of specimens that provided lower-quality sequences at the 5′ and 3′ ends.
The neighbour-joining method was used to construct a 50 % majority consensus tree of the mitochondrial 16S rDNA sequences using MEGA 5.2. Using the bootstrap method, replications were set at 2000. The substitution model selected was Kimura two-parameter (transitional and transversal substitutions included) with rates set to gamma distribution. The first 25 % of trees generated were discarded. Hypotrigona ruspolii (Magretti) (Meliponini) (DQ790441) and Hypotrigona gribodoi (Magretti) (DQ790440) were used to root the tree, and other Meliponini [H. gribodoi (Magretti) (DQ790440), Liotrigona mahafalya Brooks and Michener (DQ790442), Liotrigona madecassa (Saussure) (DQ790443), and Lisotrigona furva Engel (DQ790444)] were included as outgroup taxa. FigTree v1.4.0 (Rambaut 2012) was used to illustrate the phylogenetic tree produced.
3 Results
3.1 Definitions of the six Austroplebeia groups in this study
The Austroplebeia nests in this study were divided into six groups during the initial morphological analyses. The ‘symei’ and ‘australis’ groups resembled two varieties of Austroplebeia that differ in their extent of thoracic colour markings, and are kept in domesticated hives in Queensland: a dark variety known by beekeepers as Austroplebeia symei and a moderately marked variety known as A. australis. The ‘intermediate’ group had colour markings between ‘symei’ and ‘australis’. The ‘striped’ group was relatively large bees with lateral bands on the scutum. The ‘curved’ group was relatively small bees with lateral bands on the scutum; the male gonostylus was curved. The ‘cincta’ group was an Australian population resembling A. cincta from NG.
3.2 Head width and colour analysis of indigenous Austroplebeia groups
The level of thoracic colouration present in individual workers ranged from 0 to 20.2 % of the total thoracic area. Typical colouration scores were 1.2 % (‘symei’), 8.8 % (‘australis’), 16.7 % (‘striped’) and 20.2 % (‘curved’) (Figure 3).
When the mean HW and mean colour scores for each nest were plotted against each other, four distinct clusters became apparent (Figure 4). We named them ‘symei’ (n = 27), ‘australis’ (n = 15), ‘striped’ (n = 13) and ‘curved’ (n = 9). Nests from one location, near Kilcoy, QLD (26° 56′ S, 152° 33′ E), formed another loose cluster which we named ‘intermediate’ (n = 7). There was a significant difference in mean percentage of colour marking present on the thorax (F 4, 66 = 248.108, p < 0.001), with all groups significantly differing from each other. For HW, there was a significant difference among groups (F 4, 66 = 76.343, p < 0.001). However, there was no difference between ‘australis’ and ‘symei’ (p = 0.085), nor between the ‘intermediate’ group and ‘australis’ (p = 0.197) or ‘intermediate’ and ‘symei’ (p = 0.994).
3.3 Austroplebeia cincta morphology
The morphological characteristics of Daintree collected workers from the ‘cincta’ group matched those of specimens of A. cincta from NG. They were clearly distinct from those of indigenous Austroplebeia groups, in both colour and pilosity. The thorax of ‘cincta’ workers had yellow/cream markings on the lateral surface of the metepisternum (Figure 5a), whereas in all indigenous groups, these areas had no markings. The face of ‘cincta’ workers had distinct dull to bright yellow markings, whereas the indigenous groups had only diffused dull cream markings (Figure 5b–d).
a ‘cincta’ marking on the metepisternum, unlike the indigenous Austroplebeia groups. b ‘symei’, c ‘striped’ and d ‘cincta’ facial markings. ‘cincta’ has distinct facial markings unlike the indigenous Austroplebeia groups. Indigenous groups also have plumose hair on paraocular and supra-antennal areas.
Pilosity was quite different between the ‘cincta’ and other groups; ‘cincta’ workers’ thoracic mesepisternum and hypoepimeral areas appeared shiny with short bristles, whereas all other groups had thick plumose hairs in these areas. The ‘cincta’ workers’ faces appeared relatively shiny, as most of the hair covering was fine and sparse. In contrast, in all other groups, the faces were covered with thick white hairs including well-developed plumose hair on the paraocular and supra-antennal areas (Figure 5b–d).
3.4 Male morphology
SEM images of dissected genitalia showed that some males collected from WA and the Cobourg Peninsula NT had gonostyli which were curved at the tip (hence the group name ‘curved’), while all the other groups had straight-tipped gonostyli (Figure 6a, b). Stereomicroscope images of dissected genitalia as well as intact genitalia of pinned male specimens also showed these differences between the ‘curved’ specimens and the other groups (Figure 6c, d). It was therefore possible to separate the ‘curved’ group from the other groups—‘symei’, ‘intermediate’, ‘australis’ and ‘striped’—using distinct morphological differences in the male genitalia.
3.5 Geometric morphometry of wing venations
Six factors with eigenvalues >1 were extracted in the PCA and explained a total of 75.34 % of the total variability of the data. The first principal component explained 20.37 % of the data variability and the second principal component explained 19.70 % of the variability of the relative positions of the landmarks. The relative position of landmarks that contributed most to the first component were the Cartesian coordinate ‘y’ in the landmark 10 and the ‘x’ position in the landmarks 2, 5 and 7. The most informative positions contributing to the second component were the ‘x’ position in the landmarks 3, 4 and 10 the ‘y’ position of the landmarks 6 and 9. The ‘cincta’ group was placed mostly in the upper right-hand quadrant and the ‘curved’ was placed on the upper quadrants. The ‘striped’ group was placed in the left-hand quadrants (Figure 7). An analysis of Mahalanobis distances between the centroids indicated that there were significant differences (p < 0.005) between all groups (Table I).
The dendogram of morphological proximity (Figure 8) placed ‘cincta’ in an isolated branch, while the other four groups were split into sub-branches. The coefficient of cophenetic correlation was 0.969, indicating that the dendogram was a good two-dimensional representation of the calculated distances (Kalinowsi 2009).
The discriminant analysis using functions 1 and 2 defined the ‘cincta’ and ‘curved’ groups but superimposed the other groups, ‘australis’, ‘symei’ and ‘striped’ (Figure 9).
3.6 Molecular analysis and geographic distribution
The length of the 16S rDNA amplicons sequenced in this study varied from 416 bp, for the ‘cincta’ group, and up to 420 bp for the remaining groups.
The phylogenetic analysis (Figure 10) demonstrated that the ‘curved’ group formed the basal clade of the Austroplebeia taxa examined. Unfortunately, due to the short sequence lengths (315 bp), the clade including the ‘australis’, ‘intermediate’, ‘striped’, ‘symei’ and ‘cincta’ specimens was unresolved. The ‘australis’ specimens formed one group, with six specimens presenting identical sequences to the A. australis specimen from GenBank. The ‘intermediate’ specimens produced identical sequences and formed a sub-clade within the ‘australis’ group, resulting from a transversional single-nucleotide polymorphism (SNP). One ‘australis’ specimen (australis_Tar) fell outside this group due to one transversional SNP.
Majority consensus 16S mtDNA (315 bp) neighbour-joining phylogenetic tree for Austroplebeia specimens. The values at the nodes represent bootstrap values labelled according to their original HW/colour grouping (Figure 4).
In the dataset presented, there was only one transversional SNP difference between the QLD ‘symei’ and the QLD ‘striped’ specimens. Again, one transitional difference was found between the QLD ‘striped’ and the NT ‘striped’ groups. Notably, of the differences between the QLD ‘symei’ and the NT ‘symei’, one to two transversional and two transitional SNPs were identified. The ‘cincta’ group displayed the greatest difference within all analysed Austroplebeia specimens, by five to seven SNPs.
The morphological characteristics of HW and body coloration placed one nest (C58) within the outer margins of the ‘symei’ cluster, and within close proximity to the ‘intermediate’ group (Figure 4). However, this nest shared the genotype found in the ‘australis’ group (Figure 10).
The maximum geographic distances between the sampled nests within the various groups (Figure 11) were as follows: ‘curved’ 800 km, ‘australis’ 900 km, ‘intermediate’ 50 km, ‘symei’ 2400 km, ‘striped’ 1500 km and ‘cincta’ 1 km.
4 Discussion
The data produced during these studies clearly demonstrated that there were fewer than nine species within the genus Austroplebeia. By using multiple diagnostic methodologies, we attempted to identify different characteristics within six morphologically distinct groups: ‘symei’, ‘intermediate’, ‘australis’, ‘striped’, ‘curved’ and ‘cincta’. Four diagnostic methodologies were used and compared: (1) colour, pilosity and size of worker bees, (2) genital morphology of males, (3) morphometric analysis of wing venations and (4) molecular analysis of 16S rDNA. The molecular data were limited as it was only possible to extract a short amplicon of one gene from the old dried specimens that were available. The combined evidence from the four methodologies suggests the separation of two groups, ‘curved’ and ‘cincta’, plus an unresolved species complex.
The ‘curved’ group was definitively delimited by all analyses. The ‘cincta’ group showed clear differences in worker colour and pilosity and was also well supported by the wing morphometric analysis. In the molecular phylogeny, the placement of the ‘cincta’ group was not resolved, but it displayed the highest number of SNPs in the study. No ‘cincta’ males were available for examination of genital morphology. The ‘cincta’ group bees from Queensland could not be separated from our New Guinea A. cincta specimens by colour, pilosity or wing morphometric analysis.
The ‘australis’ group could not be separated from ‘symei’ or ‘striped’ by genital morphology and was only weakly separated by the wing morphometric and worker colour analyses. However, in the molecular phylogeny, the ‘australis’ specimens formed a group which also contained specimens from nest C58 (identified as ‘symei’ based on HW and colour) and the ‘intermediate’ nests. This suggests that ‘australis’ group may have had a wider range of thoracic colour patterns than initially proposed. The 16S rDNA of the ‘australis’ group was very conserved, with identical sequences found in nests sampled 900 km apart.
Separation of the ‘symei’ and ‘striped’ groups was well supported by the worker colour analysis. However, the wing morphometric analysis and the molecular analysis suggested that these groups were very similar and may possibly be a species complex. The relationship between QLD ‘symei’ and QLD ‘striped’ was unresolved in the molecular phylogeny. Higher numbers of SNPs were observed between the QLD ‘symei’ and NT ‘symei’ samples, which were separated by geographical distances of up to 2400 km.
The molecular differentiation between all groups except ‘curved’ and ‘cincta’ was very low for the mitochondrial 16S rDNA fragment. Perhaps, the Austroplebeia species may not yet be fully differentiated at this locus, or some low-level hybridisation between the different groups may persist resulting in mitochondrial introgression. The resolution of the molecular data may also have been diminished by the presence of nuclear mitochondrial copies of this locus that can impact phylogenetic analyses (Bensasson et al. 2001).
The current descriptions which identify the species within the genus Austroplebeia are inadequate and, as such, may hinder their identification, ecological assessment and management. Delimiting the Austroplebeia species has posed a frustrating and long-term problem (Moure 1961; Michener 1990; Rasmussen 2008), and the small amount of research using Austroplebeia spp. has been less than helpful in this area (Costa et al. 2003; Rasmussen and Cameron 2007).
The paucity of morphological differences in Austroplebeia creates great difficulties for the taxonomist. None of our methodologies were sufficient on their own to delimit the Austroplebeia species in this collection. Nevertheless, the groups delimited by the four combined methodologies have provided a useful foundation for further investigations into this genus. The species names assigned to these groups will be published separately, in a revision of the genus Austroplebeia that is in preparation (Dollin, A.E., Dollin, L.J., and Rasmussen, C., unpubl. data).
References
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410
Batley, M., Hogendoorn, K. (2009) Diversity and conservation status of native Australian bees. Apidologie 40, 347–354
Bensasson, D., Zhang, D.X., Hartl, D.L., Hewitt, G.M. (2001) Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol. Evol. 16, 314–321
Cardale, J.C. (1993) Hymenoptera: Apoidea. In: Houston, W.W.K., Maynard, G.V. (eds.) Zoological Catalogue of Australia, vol. 10, pp. 318–320. Australian Government Publishing Service, Canberra
Cockerell, T.D.A. (1905) Descriptions and records of bees. Ann. Mag. Nat. Hist. 16, 216–225
Cockerell, T.D.A. (1910) New and little-known bees. Trans. Am. Entomol. Soc. 36, 199–249
Costa, M.A., Del Lama, M.A., Melo, G.A.R., Sheppard, W.S. (2003) Molecular phylogeny of the stingless bees (Apidae, Apinae, Meliponini) inferred from mitochondrial 16S rDNA sequences. Apidologie 34, 73–84
Dias, B.S.F., Raw, A., Imperatriz-Fonseca, V.L. (1999) International pollinators initiative: The São Paulo declaration on pollinators, report on the recommendations of the workshop on the conservation and sustainable use of pollinators in agriculture with emphasis on bees. Brazilian Ministry of the Environment, Sao Paulo
Dollin, A.E., Dollin, L.J., Sakagami, S.F. (1997) Australian stingless bees of the genus Trigona (Hymenoptera: Apidae). Invertebr. Taxon. 11, 861–896
Francoy, T.M., Wittmann, D., Drauschke, M., Müller, S., Steinhage, V., Bezerra-Laure, M.A.F., Jong, D.D., Gonçalves, L.S. (2008) Identification of Africanized honey bees through wing morphometrics: two fast and efficient procedures. Apidologie 39, 488–494
Francoy, T.M., Grassi, M., Imperatriz-Fonseca, V., de Jesús May-Itzá, W., Quezada-Euán, J. (2011) Geometric morphometrics of the wing as a tool for assigning genetic lineages and geographic origin to Melipona beecheii (Hymenoptera: Meliponini). Apidologie 42, 499–507
Friese, H. (1898) Die Trigona-Arten Australiens (The Australian Trigona). Termeszet Füzetek (World Nat Pap) 21, 427–431
Imperatriz-Fonseca, V.L. (2010) Biodiversidade, conservação e uso sustentado De polinizadores, com ênfase em abelhas, no Brasil (Biodiversity, conservation and sustainable use of pollinators, mainly bees). In: Simões, Z.L.P., Marco Antonio, D.S., Bitondi, M.M.G. (eds.) Anais do IX Encontro sobre Abelhas: Genética e Biologia Evolutiva de Abelhas Dados Internacionais de Catalogação na Publicação, pp. 698. Ribeirão Preto, Brazil
Kalinowsi, S.T. (2009) How well do evolutionary trees describe genetic relationships between populations? Heredity 102, 506–513
Kumar, S., Filipski, A. (2007) Multiple sequence alignment: In pursuit of homologous DNA positions. Genome Res. 17, 127–135
Michener, C.D. (1990) Classification of the Apidae (Hymenoptera). Univ. Kansas Sci. Bull. 54, 75–164
Michener, C.D. (2000) The Bees of the World. Johns Hopkins University Press, Baltimore, USA
Moure, J.S. (1961) A preliminary supra-specific classification of the old world Meliponine bees (Hymenoptera, Apoidea). Stud. Entomol. 4, 181–242
Rambaut, A. (2012) FigTree v1.4.0. http://tree.bio.ed.ac.uk/software/figtree/ Accessed March 2013
Rasmussen, C. (2008) Catalog of the Indo-Malayan/Australasian stingless bees (Hymenoptera: Apidae: Meliponini). Zootaxa 1935, 1–80
Rasmussen, C., Cameron, S. (2007) A molecular phylogeny of the old world stingless bees (Hymenoptera: Apidae: Meliponini) and the non-monophyly of the large genus Trigona. Syst. Entomol. 32, 26–39
Rayment, T. (1932) The stingless bees of Australia: 6. The finding of a new species. Vic. Nat. 49, 104–107
Rohlf, F.J. (2005) tpsDig2, version 2.04, Department of Ecology and Evolution, State University of New York. Stony Brook, New York
Saitou, N., Nei, M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425
StatSoft, I. (2001) STATISTICA (data analysis software system), version 6. Available at: www.statsoft.com. Accessed October 2010
Tamura, K., Dudley, J., Nei, M., Kumar, S. (2007) MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011) Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739
Vogler, A.P., Monaghan, M.T. (2007) Recent advances in DNA taxonomy. J. Zool. Syst. Evol. Res. 45, 1–10
Acknowledgments
Our thanks go to Les Dollin, Australian Native Bee Research Centre (www.aussiebee.com.au), for his skills and enthusiasm in locating the native bee nests; to Rute Brito, Universidade Federal de Uberlândia, for alerting us to the existence of the ‘cincta’ bees and to naturalist Lewis Roberts, for collecting ‘cincta’ specimens from the Daintree area, northern QLD; and to Jen Morrow, Aiman Jajo, Paul Smith, Adriyan Milev and Liz Kabanoff, University of Western Sydney, and Michael Batley, Australian Museum, for their technical support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Manuscript editor: Marina Meixner
Délimitation des espèces à l'intérieur du genre Austroplebeia , une abeille sans aiguillon d'Australie, par l'utilisation de différentes méthodologies
coloration thoracique / morphométrie géométrique / ADNr 16S / génitalia mâles / Meliponini / pilosité
Abgrenzung von Arten innerhalb der Gattung Austroplebeia, einer stachellosen Biene aus Australien, mit verschiedenen Methoden
Thoraxfärbung / geometrische Morphometrie / mitochondriale 16S rDNA / männliche Genitalien / Meliponini / Behaarung
To the memory of Dr. Shôichi F. Sakagami, Professor Emeritus of Hokkaido University.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 442 kb)
Rights and permissions
About this article
Cite this article
Halcroft, M.T., Dollin, A., Francoy, T.M. et al. Delimiting the species within the genus Austroplebeia, an Australian stingless bee, using multiple methodologies. Apidologie 47, 76–89 (2016). https://doi.org/10.1007/s13592-015-0377-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-015-0377-7