Abstract
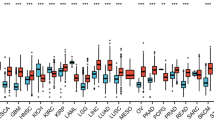
LINC00941, also known as lncRNA-MUF, is an intergenic non-coding RNA located on chromosome 12p11.21. It actively participates in a complex competing endogenous RNA network, regulating the expression of microRNA and its downstream proteins. Through transcriptional and post-transcriptional regulation, LINC00941 plays a vital role in multiple signaling pathways, influencing cell behaviors such as tumor cell proliferation, epithelial–mesenchymal transition, migration, and invasion. Noteworthy is its consistently high expression in various tumor types, closely correlating with clinicopathological features and cancer prognoses. Elevated LINC00941 levels are associated with adverse clinical outcomes, including increased tumor size, extensive lymphatic metastasis, and distant metastasis, leading to poorer survival rates across different cancers. Additionally, LINC00941 and its associated genes are linked to various targeted drugs available in the market. In this comprehensive review, we systematically summarize existing studies, detailing LINC00941's differential expression, clinicopathological and prognostic implications, regulatory mechanisms, and associated therapeutic drugs. Our analysis includes relevant charts and incorporates bioinformatics analyses to verify LINC00941's differential expression in pan-cancer and explore potential transcriptional regulation patterns of downstream targets. This work not only establishes a robust data foundation but also guides future research directions. Given its potential as a significant cancer biomarker and therapeutic target, further investigation into LINC00941's differential expression and regulatory mechanisms is essential.





Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- ACC:
-
Adrenocortical carcinoma
- ALL:
-
Acute lymphoblastic leukemia
- AML:
-
Acute myeloid leukemia
- BLCA:
-
Bladder urothelial carcinoma
- BRCA:
-
Breast invasive carcinoma
- ceRNA:
-
Competing endogenous RNA
- CESC:
-
Cervical esophageal squamous cell carcinoma
- CHOL:
-
Cholangiocarcinoma
- COAD:
-
Colon adenocarcinoma
- CRC:
-
Colorectal cancer
- DFS:
-
Disease-free survival
- EMT:
-
Epithelial–mesenchymal transition
- ESCA:
-
Esophageal carcinoma
- ESCC:
-
Esophageal squamous cell carcinoma
- FAK:
-
Focal adhesion kinase
- GBM:
-
Glioblastoma multiforme
- GC:
-
Gastric cancer
- HCC:
-
Hepatocellular carcinoma
- HNSC:
-
Head and neck squamous cell carcinoma
- KICH:
-
Kidney chromophobe
- KIRC:
-
Kidney renal clear cell carcinoma
- KIRP:
-
Kidney papillary cell carcinoma
- LATS1/2:
-
Large tumor suppressor 1/2
- LC:
-
Laryngocarcinoma
- LGG:
-
Lower grade glioma
- LIHC:
-
Liver hepatocellular carcinoma
- LncRNA:
-
Long non-coding RNA
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- miRNA:
-
MicroRNA
- miRNA-RISC:
-
MiRNA-containing RNA-induced silencing complex
- MREs:
-
MicroRNA response elements
- MST1/2:
-
Mammalian sterile20-like kinase 1/2
- NBL:
-
Neuroblastoma
- NSCLC:
-
Non-small cell lung cancerr
- OV:
-
Ovarian cancer
- OS:
-
Overall survival
- OSCC:
-
Oral squamous cell carcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- PC:
-
Pancreatic cancer
- PCPG:
-
Pheochromocytoma and paraganglioma cancer
- PDAC:
-
Pancreatic ductal adenocarcinoma
- PP2A:
-
Protein phosphatase 2A
- PRAD:
-
Prostate adenocarcinoma
- PTC:
-
Primes papillary thyroid cancer
- READ:
-
Rectum adenocarcinoma
- SARC:
-
Sarcoma
- SKCM:
-
Skin cutaneous melanoma
- SPP:
-
Signaling pathways project
- STAD:
-
Stomach adenocarcinoma
- TGCT:
-
Testicular germ cell tumors
- TGF- β:
-
Transforming growth factor-β
- THCA:
-
Thyroid carcinoma
- THYM:
-
Thymoma
- TNM:
-
Tumor node metastasis
- TSS:
-
Transcription start site
- UCEC:
-
Endometrioid cancer
- UCS:
-
Uterine carcinosarcoma
- UCSC Genome Browser:
- CADDIE database:
- ChIP-Atlas:
- JASPAR 2022 database:
- SPP database:
- TCGA database:
References
Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65:625–39.
Li C, Ni YQ, Xu H, et al. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct Target Ther. 2021;6:383.
Ai Y, Wu S, Zou C, Wei H. LINC00941 promotes oral squamous cell carcinoma progression via activating CAPRIN2 and canonical WNT/beta-catenin signaling pathway. J Cell Mol Med. 2020;24:10512–24.
Zhang Q, Shen J, Wu Y, Ruan W, Zhu F, Duan S. LINC00520: A Potential Diagnostic and Prognostic Biomarker in Cancer. Front Immunol. 2022;13:845418.
Bolha L, Ravnik-Glavac M, Glavac D. Long Noncoding RNAs as Biomarkers in Cancer. Dis Markers. 2017;2017:7243968.
Zhang Q, Zhong C, Duan S. The tumorigenic function of LINC00858 in cancer. Biomed Pharmacother. 2021;143:112235.
Zhang Y, Zhu H, Sun N, et al. Linc00941 regulates esophageal squamous cell carcinoma via functioning as a competing endogenous RNA for miR-877-3p to modulate PMEPA1 expression. Aging (Albany NY). 2021;13:17830–46.
Yan X, Zhang D, Wu W, et al. Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res. 2017;77:6704–16.
Morgenstern E, Kretz M. The human long non-coding RNA LINC00941 and its modes of action in health and disease. Biol Chem. 2023. https://doi.org/10.1515/hsz-2023-0183.
Kunej T, Obsteter J, Pogacar Z, Horvat S, Calin GA. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 2014;51:344–57.
Wang J, He Z, Liu X, et al. LINC00941 promotes pancreatic cancer malignancy by interacting with ANXA2 and suppressing NEDD4L-mediated degradation of ANXA2. Cell Death Dis. 2022;13:718.
Li Z, Jin Q, Sun Y. LINC00941 promoted in vitro progression and glycolysis of laryngocarcinoma by upregulating PKM via activating the PI3K/AKT/mTOR signaling pathway. J Clin Lab Anal. 2022;36:e24406.
Shree B, Tripathi S, Sharma V. Transforming Growth Factor-Beta-Regulated LncRNA-MUF Promotes Invasion by Modulating the miR-34a Snail1 Axis in Glioblastoma Multiforme. Front Oncol. 2021;11:788755.
Wu N, Jiang M, Liu H, et al. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-beta/SMAD2/3 signaling pathway. Cell Death Differ. 2021;28:219–32.
Liu J, Li Z, Zhang T, et al. Long Noncoding RNA LINC00941 Promotes Cell Proliferation and Invasion by Interacting with hnRNPK in Oral Squamous Cell Carcinoma. Nutr Cancer. 2022;74:2983–95.
Wang J, He Z, Xu J, Chen P, Jiang J. Long noncoding RNA LINC00941 promotes pancreatic cancer progression by competitively binding miR-335-5p to regulate ROCK1-mediated LIMK1/Cofilin-1 signaling. Cell Death Dis. 2021;12:36.
Xu M, Cui R, Ye L, et al. LINC00941 promotes glycolysis in pancreatic cancer by modulating the Hippo pathway. Mol Ther Nucleic Acids. 2021;26:280–94.
Lu JT, Yan ZY, Xu TX, et al. Reciprocal regulation of LINC00941 and SOX2 promotes progression of esophageal squamous cell carcinoma. Cell Death Dis. 2023;14:72.
Luo C, Tao Y, Zhang Y, et al. Regulatory network analysis of high expressed long non-coding RNA LINC00941 in gastric cancer. Gene. 2018;662:103–9.
Beeraka NM, Gu H, Xue N, et al. Testing lncRNAs signature as clinical stage-related prognostic markers in gastric cancer progression using TCGA database. Exp Biol Med (Maywood). 2022;247:658–71.
Fang Y, Yang Y, Zhang X, et al. A Co-Expression Network Reveals the Potential Regulatory Mechanism of lncRNAs in Relapsed Hepatocellular Carcinoma. Front Oncol. 2021;11:745166.
Chang L, Zhou D, Luo S. Novel lncRNA LINC00941 Promotes Proliferation and Invasion of Colon Cancer Through Activation of MYC. Onco Targets Ther. 2021;14:1173–86.
Ren MH, Chen S, Wang LG, Rui WX, Li P. LINC00941 Promotes Progression of Non-Small Cell Lung Cancer by Sponging miR-877-3p to Regulate VEGFA Expression. Front Oncol. 2021;11:650037.
Fang L, Wang SH, Cui YG, Huang L. LINC00941 promotes proliferation and metastasis of pancreatic adenocarcinoma by competitively binding miR-873-3p and thus upregulates ATXN2. Eur Rev Med Pharmacol Sci. 2021;25:1861–8.
Chan JJ, Tay Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int J Mol Sci. 2018;19:1310.
Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). 2021;41:109–20.
Lupianez DG, Kraft K, Heinrich V, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–25.
Debaugny RE, Skok JA. CTCF and CTCFL in cancer. Curr Opin Genet Dev. 2020;61:44–52.
Li Y, Haarhuis JHI, Sedeno Cacciatore A, et al. The structural basis for cohesin-CTCF-anchored loops. Nature. 2020;578:472–6.
Fang C, Wang Z, Han C, et al. Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation. Genome Biol. 2020;21:247.
Lu J, Yu L, Xie N, Wu Y, Li B. METTL14 Facilitates the Metastasis of Pancreatic Carcinoma by Stabilizing LINC00941 in an m6A-IGF2BP2-Dependent Manner. J Cancer. 2023;14:1117–31.
Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46.
Becnel LB, Ochsner SA, Darlington YF, et al. Discovering relationships between nuclear receptor signaling pathways, genes, and tissues in Transcriptomine. Sci Signal. 2017. https://doi.org/10.1126/scisignal.aah6275.
Zou Z, Ohta T, Miura F, Oki S. ChIP-Atlas 2021 update: a data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res. 2022;50:W175–82.
Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50:D165–73.
Karreth FA, Pandolfi pp. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi pp. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8.
Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–7.
Cheng SC, Quintin J, Cramer RA, et al. mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684.
Sun Q, Chen X, Ma J, et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc Natl Acad Sci U S A. 2011;108:4129–34.
Tian T, Li X, Zhang J. mTOR Signaling in Cancer and mTOR Inhibitors in Solid Tumor Targeting Therapy. Int J Mol Sci. 2019;20:755.
Molli PR, Li DQ, Murray BW, Rayala SK, Kumar R. PAK signaling in oncogenesis. Oncogene. 2009;28:2545–55.
Dawson JC, Serrels A, Stupack DG, Schlaepfer DD, Frame MC. Targeting FAK in anticancer combination therapies. Nat Rev Cancer. 2021;21:313–24.
Katsuno M, Adachi H, Banno H, Suzuki K, Tanaka F, Sobue G. Transforming growth factor-beta signaling in motor neuron diseases. Curr Mol Med. 2011;11:48–56.
Xie F, Ling L, van Dam H, Zhou F, Zhang L. TGF-beta signaling in cancer metastasis. Acta Biochim Biophys Sin (Shanghai). 2018;50:121–32.
Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nat Rev Drug Discov. 2012;11:790–811.
Heldin CH, Moustakas A. Signaling Receptors for TGF-beta Family Members. Cold Spring Harb Perspect Biol. 2016;8:a022053.
Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–30.
Muralidhar S, Filia A, Nsengimana J, et al. Vitamin D-VDR Signaling Inhibits Wnt/beta-Catenin-Mediated Melanoma Progression and Promotes Antitumor Immunity. Cancer Res. 2019;79:5986–98.
Yu F, Yu C, Li F, et al. Wnt/beta-catenin signaling in cancers and targeted therapies. Signal Transduct Target Ther. 2021;6:307.
Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–50.
Lee MH, Kundu JK, Chae JI, Shim JH. Targeting ROCK/LIMK/cofilin signaling pathway in cancer. Arch Pharm Res. 2019;42:481–91.
Lu G, Zhou Y, Zhang C, Zhang Y. Upregulation of LIMK1 Is Correlated With Poor Prognosis and Immune Infiltrates in Lung Adenocarcinoma. Front Genet. 2021;12:671585.
Ishaq M, Lin BR, Bosche M, et al. LIM kinase 1 - dependent cofilin 1 pathway and actin dynamics mediate nuclear retinoid receptor function in T lymphocytes. BMC Mol Biol. 2011;12:41.
Dey A, Varelas X, Guan KL. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat Rev Drug Discov. 2020;19:480–94.
Ma S, Meng Z, Chen R, Guan KL. The Hippo Pathway: Biology and Pathophysiology. Annu Rev Biochem. 2019;88:577–604.
Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118.
Liu L, Michowski W, Kolodziejczyk A, Sicinski P. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat Cell Biol. 2019;21:1060–7.
Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495–516.
Xu X, Lai Y, Hua ZC. Apoptosis and apoptotic body: disease message and therapeutic target potentials. Biosci Rep. 2019;39:495.
Babaei G, Aziz SG, Jaghi NZZ. EMT, cancer stem cells and autophagy; The three main axes of metastasis. Biomed Pharmacother. 2021;133:110909.
Lusby R, Dunne P, Tiwari VK. Tumour invasion and dissemination. Biochem Soc Trans. 2022;50:1245–57.
Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20:629–51.
Shao J, Zhang B, Kuai L, Li Q. Integrated analysis of hypoxia-associated lncRNA signature to predict prognosis and immune microenvironment of lung adenocarcinoma patients. Bioengineered. 2021;12:6186–200.
Chen H, Zhou C, Hu Z, et al. Construction of an algorithm based on oncosis-related LncRNAs comprising the molecular subtypes and a risk assessment model in lung adenocarcinoma. J Clin Lab Anal. 2022;36:e24461.
Gugnoni M, Manicardi V, Torricelli F, et al. Linc00941 Is a Novel Transforming Growth Factor beta Target That Primes Papillary Thyroid Cancer Metastatic Behavior by Regulating the Expression of Cadherin 6. Thyroid. 2021;31:247–63.
Zou Z, Ma T, He X, et al. Long intergenic non-coding RNA 00324 promotes gastric cancer cell proliferation via binding with HuR and stabilizing FAM83B expression. Cell Death Dis. 2018;9:717.
Wang S, Cheng Y, Yang P, Qin G. Silencing of Long Noncoding RNA LINC00324 Interacts with MicroRNA-3200-5p to Attenuate the Tumorigenesis of Gastric Cancer via Regulating BCAT1. Gastroenterol Res Pract. 2020;2020:4159298.
Liu H, Wu N, Zhang Z, et al. Long Non-coding RNA LINC00941 as a Potential Biomarker Promotes the Proliferation and Metastasis of Gastric Cancer. Front Genet. 2019;10:5.
Hartung M, Anastasi E, Mamdouh ZM, et al. Cancer driver drug interaction explorer. Nucleic Acids Res. 2022;50:W138–44.
Wang Y, Zhao D, Wang H, et al. Long non-coding RNA-LINC00941 promotes the proliferation and invasiveness of glioma cells. Neurosci Lett. 2023;795:136964.
Pan H, Wei W, Fu G, Pan J, Jin B. LINC00941 Promotes Cell Malignant Behavior and Is One of Five Costimulatory Molecule-Related lncRNAs That Predict Prognosis in Renal Clear Cell Carcinoma. Medicina (Kaunas). 2023;59:187.
Funding
This study was supported by the Qiantang Scholars Fund in Hangzhou City University (No. 210000–581835).
Author information
Authors and Affiliations
Contributions
QY, XS, YC, ZW, WH, QX, and YM collected and analyzed the literature, drafted the figures, and wrote the manuscript. SD and HL conceived the idea and gave the final approval of the submitted version. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare no competing interests.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yan, Q., Su, X., Chen, Y. et al. LINC00941: a novel player involved in the progression of human cancers. Human Cell 37, 167–180 (2024). https://doi.org/10.1007/s13577-023-01002-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-023-01002-5