Abstract
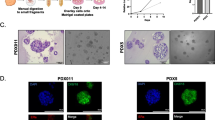
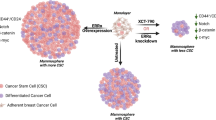
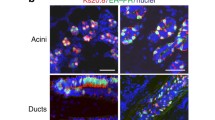
Estrogen receptor (ER) expression in breast cancer can change during progression and the treatment, but the mechanism has not been well studied. In this study, we successfully prepared organoids from samples obtained from 33 luminal-type breast cancer patients and studied their ER expression. The expression status was well maintained in primary organoids, whereas it decreased after passaging in most of the cases. In fact, the studied organoid lines were classified into those that retained a high level of ER expression (9%), those that completely lost it (9%), and those that repressed it to varying degrees (82%). In some cases, the ER expression was suddenly and drastically decreased after passaging. Marker protein immunohistochemistry revealed that after passaging, the differentiation status shifted from a luminal- to a basal-like status. Differentially expressed genes suggested the activation of NOTCH signaling in the passaged organoids, wherein a NOTCH inhibitor was able to substantially rescue the decreased ER expression and alter the differentiation status. Our findings suggest that the differentiation status of luminal-type cancer cells is quite flexible, and that by inhibiting the NOTCH signaling we can preserve the differentiation status of luminal-type breast cancer organoids.






Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are available in DDBJ (accession number: DRR412393-DRR412396).
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
Clark GM, Osborne CK, McGuire WL. Correlations between estrogen receptor, progesterone receptor, and patient characteristics in human breast cancer. J Clin Oncol. 1984;2(10):1102–9.
Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–47.
Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet. 2015;386(10001):1341–1352.
Aitken SJ, Thomas JS, Langdon SP, Harrison DJ, Faratian D. Quantitative analysis of changes in ER, PR and HER2 expression in primary breast cancer and paired nodal metastases. Ann Oncol. 2010;21(6):1254–61.
Lindstrom LS, Karlsson E, Wilking UM, Johansson U, Hartman J, Lidbrink EK, Hatschek T, Skoog L, Bergh J. Clinically used breast cancer markers such as estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J Clin Oncol. 2012;30(21):2601–8.
Herrera-Abreu MT, Palafox M, Asghar U, Rivas MA, Cutts RJ, Garcia-Murillas I, Pearson A, Guzman M, Rodriguez O, Grueso J, et al. Early adaptation and acquired resistance to CDK4/6 inhibition in Estrogen receptor-positive breast cancer. Cancer Res. 2016;76(8):2301–13.
Comsa S, Cimpean AM, Raica M. The story of MCF-7 breast cancer cell line: 40 years of experience in research. Anticancer Res. 2015;35(6):3147–54.
Burdall SE, Hanby AM, Lansdown MR, Speirs V. Breast cancer cell lines: friend or foe? Breast Cancer Res. 2003;5(2):89–95.
Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4(4):359–65.
Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172(1–2):373-386 e310.
Dekkers JF, van Vliet EJ, Sachs N, Rosenbluth JM, Kopper O, Rebel HG, Wehrens EJ, Piani C, Visvader JE, Verissimo CS, et al. Long-term culture, genetic manipulation and xenotransplantation of human normal and breast cancer organoids. Nat Protoc. 2021;16(4):1936–65.
Campaner E, Zannini A, Santorsola M, Bonazza D, Bottin C, Cancila V, Tripodo C, Bortul M, Zanconati F, Schoeftner S, et al. Breast cancer organoids model patient-specific response to drug treatment. Cancers (Basel). 2020;12(12):3869.
Pan B, Li X, Zhao D, Li N, Wang K, Li M, Zhao Z. Optimizing individualized treatment strategy based on breast cancer organoid model. Clin Transl Med. 2021;11(4): e380.
Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18(7):407–18.
Kondo J, Endo H, Okuyama H, Ishikawa O, Iishi H, Tsujii M, Ohue M, Inoue M. Retaining cell-cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc Natl Acad Sci USA. 2011;108(15):6235–40.
Rosenbluth JM, Schackmann RCJ, Gray GK, Selfors LM, Li CM, Boedicker M, Kuiken HJ, Richardson A, Brock J, Garber J, et al. Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat Commun. 2020;11(1):1711.
Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, Cervera N, Fekairi S, Xerri L, Jacquemier J, Birnbaum D, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25(15):2273–84.
Eroles P, Bosch A, Perez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38(6):698–707.
Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10(1):R10.
Lv Y, Wang X, Li X, Xu G, Bai Y, Wu J, Piao Y, Shi Y, Xiang R, Wang L. Nucleotide de novo synthesis increases breast cancer stemness and metastasis via cGMP-PKG-MAPK signaling pathway. PLoS Biol. 2020;18(11): e3000872.
Guillen KP, Fujita M, Butterfield AJ, Scherer SD, Bailey MH, Chu Z, DeRose YS, Zhao L, Cortes-Sanchez E, Yang CH, et al. A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology. Nat Cancer. 2022;3(2):232–50.
Haughian JM, Pinto MP, Harrell JC, Bliesner BS, Joensuu KM, Dye WW, Sartorius CA, Tan AC, Heikkila P, Perou CM, et al. Maintenance of hormone responsiveness in luminal breast cancers by suppression of Notch. Proc Natl Acad Sci USA. 2012;109(8):2742–7.
Nikulin SV, Alekseev BY, Sergeeva NS, Karalkin PA, Nezhurina EK, Kirsanova VA, Sviridova IK, Akhmedova SA, Volchenko NN, Bolotina LV, et al. Breast cancer organoid model allowed to reveal potentially beneficial combinations of 3,3’-diindolylmethane and chemotherapy drugs. Biochimie. 2020;179:217–27.
Whittle JR, Vaillant F, Surgenor E, Policheni AN, Giner G, Capaldo BD, Chen HR, Liu HK, Dekkers JF, Sachs N, et al. Dual targeting of CDK4/6 and BCL2 pathways augments tumor response in Estrogen receptor-positive breast cancer. Clin Cancer Res. 2020;26(15):4120–34.
Jin X, Ge LP, Li DQ, Shao ZM, Di GH, Xu XE, Jiang YZ. LncRNA TROJAN promotes proliferation and resistance to CDK4/6 inhibitor via CDK2 transcriptional activation in ER+ breast cancer. Mol Cancer. 2020;19(1):87.
Piulats JM, Kondo J, Endo H, Ono H, Hagihara T, Okuyama H, Nishizawa Y, Tomita Y, Ohue M, Okita K, et al. Promotion of malignant phenotype after disruption of the three-dimensional structure of cultured spheroids from colorectal cancer. Oncotarget. 2018;9(22):15968–83.
Kretzschmar K, Clevers H. Organoids: modeling development and the stem cell niche in a dish. Dev Cell. 2016;38(6):590–600.
Rand MD, Grimm LM, Artavanis-Tsakonas S, Patriub V, Blacklow SC, Sklar J, Aster JC. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20(5):1825–35.
Liu W, Morgan KM, Pine SR. Activation of the Notch1 stem cell signaling pathway during routine cell line subculture. Front Oncol. 2014;4:211.
Okuyama H, Kondo J, Sato Y, Endo H, Nakajima A, Piulats JM, Tomita Y, Fujiwara T, Itoh Y, Mizoguchi A, et al. Dynamic change of polarity in primary cultured spheroids of human colorectal adenocarcinoma and its role in metastasis. Am J Pathol. 2016;186(4):899–911.
Acknowledgements
We thank Mr. Hiromitsu Tazawa (Clinical Bio-resource Center of Kyoto University’s Hospital) for his administrative support and Mitsuaki Ishida for his support in the pathological analysis.
Funding
This research was supported by a collaboration grant from Kyoto University-KBBM (H.U., C.S., J.K., Y.M., M.M., T.S., M.I,) and the Takeda Science Foundation (M.I.).
Author information
Authors and Affiliations
Contributions
MM, YM, TS, and MI designed the study. HU and CS performed the experiments. JK and RO supervised the experiments. YK, MT, and TS acquired the clinical samples and data. HU, RO, and MI conducted the data analysis. HU and MI wrote the manuscript. HU, JK, KO, RO, and TS reviewed and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
J.K., K.O., and M.I. are members of the Department of Clinical Bio-resource Research and Development at Kyoto University, which is sponsored by KBBM, Inc. H.U. and C.S. are employees of KBBM Inc. The other authors declare no conflict of interest. This work was supported in part by the collaboration grant from KBBM-Kyoto University.
Ethics approval and consent to participate
This study was approved by the institutional ethics committees of Kyoto University (R2220, R2444) and Kansai Medical University (2019245, 2021090). Written informed consent was obtained from participants. This study was performed in accordance with the Declaration of Helsinki.
Consent for publication
Written consent for publication was obtained from participants.
Informed consent
Written consent for publication was obtained from participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Uematsu, H., Saito, C., Kondo, J. et al. De-differentiation in cultures of organoids from luminal-type breast cancer is restored by inhibition of NOTCH signaling. Human Cell 36, 2099–2112 (2023). https://doi.org/10.1007/s13577-023-00975-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-023-00975-7