Abstract
Plants represent a rich repository of taxonomically restricted, yet chemically diverse, secondary metabolites that are synthesised via specific metabolic pathways. Enzyme specificity and biosynthetic gene clustering are the bottleneck of secondary metabolite evolution. As economically important food crops, grape, strawberry, and olive produce many pharmaceutically important molecules; however, their specific biosynthetic pathways remain inaccessible. Our genomic-based analysis of these three species reveal the biosynthetic diversity of their specialised secondary metabolites. We found over 20 BGCs predicted, most of which were characterised in two species, grape and strawberry. Gene annotation of the biosynthetic candidate genes predicted the production of many medically and industrially important compounds including cycloartenol, nerolidol, farnesene and valencene. Although most of the predicted clusters are concentrated in specific genomic positions, some have shown gene duplications in their clusters, which is suggestive of pseudogenes or misassembled genomes. Our genome mining and putative functional analysis of the biosynthetic genes annotated in the three species indicated the evolutionary processes that have shaped their current genetic structure and the structural diversity of their chemical compositions. Revealing the biogenetic background of these natural molecules is a step forward towards the expansion of their chemical diversification via engineering their biosynthetic genes heterologously, as well as the identification of their role in the interaction between those plants and their biotic and abiotic conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Terpenoid compounds (terpenes and molecules with a terpene moiety and additional functional groups that usually contain oxygen) are among the most diversified ancient superfamilies of natural products that emerged at the very beginning of cellular life. The presence of terpenoid-like compounds e.g. polyprenols, sterols and steranes in the cellular membrane supports this assertion (Kalua and Boss 2010; Ilc et al 2016). These ancient terpenes diversified into the ubiquitous class of small molecules that we see today, spanning over 80,000 distinct compounds across all cellular life including archaea, fungi, bacteria, plants and insects, of which plants are the lead producers (Davis and Croteau 2000; Christianson 2008; Oldfield and Lin 2012; Dellas et al 2013; Karunanithi and Zerbe 2019; Jin et al 2020;). Apart from their essential roles in plant fitness (e.g. growth and development), terpenes play a key role in defences against insect pests, herbivores, and microbial pathogens, as well as attracting pollinators and facilitating plant-plant interactions (Bohlmann and Keeling 2008; Agrawal and Heil 2012; Schmetz et al. 2014; Vaughan et al 2015; Al-Salihi and Alberti 2021). Due to their bioactivities, terpenes have been widely exploited in the production of valuable materials including biopolymers, flavours, fragrances, cosmetics, biofuels, and pharmaceuticals (Paddon et al 2013; Pateraki et al 2015; Jabir et al 2018; Booth and Bohlmann 2019). This economic relevance of terpenes has inspired scientists to further investigate them and elucidate their synthesis mechanisms and the enzymes involved in their formation, and to conclude that terpenes are generally synthesised through the conversion of the five-carbon precursor (isoprenoid) into diverse chemical derivatives, which are then functionally modified to produce the wide range of specialized terpene molecules (Wildereman and Peters 2007; Zerbe et al 2015). These reactions are mostly initiated by a distinct terpene synthase and one of the cytochrome P450 monooxygenases (Banerjee and Hamberger 2018; Bathe and Tissier 2019).
The recent ground-breaking advances in genomic and computational tools have enabled the unprecedented characterisation of terpenoid bioproducts in many plant, bacterial, and fungal species, which can be further modified via metabolic pathway engineering tools, to ultimately generate a broader spectrum of biologically active terpenes (Kumar et al 2016a, b; Jia et al 2019). Yet this generation requires the design of simple, reproducible, synthetic pathways that could minimize the number of reaction steps for the creation of scalable and cost-effective routes that can be utilised for the production of medicinally valuable natural products such as terpenes (Al-Salihi et al 2019). Genome mining approaches, for example, are useful techniques to initiate and manipulate the production of pharmaceutically and industrially important molecules. This article focuses on the potential of three economically important fruit crops, grape (Vitis vinifera), strawberry (Fragaria vesca), and olive (Olea europaea), as a source of terpenes with potential medical importance. Three species from three taxonomically distantly related families (Rosaceae, Vitaceae and Oleaceae) were chosen in order to represent the breadth of terpene molecules identified.
Vitis, or grapevine, is a genus of over 70 species, of which Vitis labrusca and V. vinifera are the most common. Historically, they were mainly cultivated in the northern hemisphere and temperate zones then moved through India into East Asia, and now are cultivated on almost all continents where the environment is suitable, accounting for 9% of all fruit production worldwide (This et al 2006; Nassiri-Asl 2016; FAO 2021). Grapes can be used as fresh fruits or can be processed to produce post-harvest products including raisins, grape juice, and wine. The chemical profile of the grape plant consists of two types of compounds, volatile and non-volatile. The volatile compounds are mainly of terpene derivatives such as mono, sesqui and triterpenes, while the non-volatile groups consist of phenylpropanoid, methoxypyrazines and the volatile sulphur thiol. The function of those compounds depends on their position in the plant: they could be a source of nutrients, cell modification requirements, or used in defence against herbivores (Dunlevy et al 2013; Parker et al 2017; Lin et al 2019). Over 50 monoterpene-like precursors such as linalool, geraniol, citronellol, terpineol and nerol have been identified from grape, with linalool derivatives being the major component. Generally, ripening berries are the richest part of the plant for terpene-like compounds. Depending on its terpene profile, grapevine can be divided into three groups: neutral (e.g. Chardonnay), moderate (e.g. Auxerrois) and high (e.g. Muscat of Alexandria) terpene concentration (Matarese et al 2014; Black et al 2015; Ilc et al 2016). Terpenes can be produced in grapevine through two pathways: the predominant or the plastidial pathway, which is responsible for the production of both monoterpene and diterpene compounds, and the primary or the mevalonate pathway, which is responsible for the production of sesquiterpene compounds (Bohlmann and Keeling 2008).
Strawberry is another economically fruit of which around 9 million tonnes are produced annually worldwide, mostly of the hybrid species Fragaria × ananassa (FAO 2021). Over 24 species of wild strawberry have been characterised. They are naturally distributed in America, Europe and Asia, of which two species Fragaria chiloensis and Fragaria virginiana were thought to have been crossbred to produce the most common cultivated species, F. x ananassa (Potter et al 2000; Bolger et al 2014); however, a recent study suggested that current cultivated strawberries are the result of the hybridisation of four species: F. viridis, F. vesca, F. iinumae, and F. nipponica (Edger et al 2019). It is included within the most fragile fruits list, due to its moist nature which makes it susceptible to water loss, external mechanical damage, and microbial contamination. It is therefore important to select or design a biological system for such fruits, to help in the prolong their shelf life, as well as their content of pharmaceutically valuable organic molecules (Perkins-Veazie and Huber 1988; Sanz et al 1999).
Strawberry’s main constituents include carbohydrates such as sucrose, fructose and glucose, and organic acids such as malic, citric acid, and volatile molecules. Strawberry also has a rich array of minerals and vitamins, such as vitamin E, vitamin C, potassium, and flavonoids to name a few (Peretto et al 2014; Skrovankova et al 2015; FAO 2021). Strawberry’s aroma profile is the result of the presence of hundreds of medicinally valuable volatile compounds including alpha linolenic acid, linoleic acid, oleic acid, quercetin, alpha terpineol, ellagic acid, gallic acid, quercitrin, linalool, ascorbic acid as well as anthocyanins. Factors like climate, the cultivar and cultivation conditions can largely influence the final concentration of the chemical constituent of strawberry plants. In a similar manner to grapevine, strawberry terpene compounds are synthesised via the plastidial and the mevalonate pathways, through which the building blocks isopentenyl diphosphate and dimethylallyl diphosphate are formed to then initiate the terpenoid condensation reactions (González-Domínguez et al 2020).
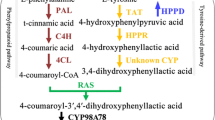
In terms of the olive plant, the O. europaea species is the only genus of the Oleaceae family that produces edible fruits and can survive harsh environmental conditions such as drought and poor soils. Around 12 million hectares of olive groves are cultivated annually (FAO 2021). In addition to its productive culture and nutritional benefits, olive plants and their by-products (mainly olive fruits and olive oil), represent a promising source of biologically active compounds for example, oleuropein and secoiridoids. Secoiridoids are a class of monoterpenes with a distinguished 3, 4- dihydropyran backbone of specialised compounds (e.g. oleosides, secoiridoids and oleosidic), that possess exocyclic functional olefinic and tyrosine groups. Despite their low yield, those molecules play a key role in determining the olive oil quality, especially its taste or bitterness as well as its medicinal properties. Oleuropein on the other hand, represents a promising antioxidant, anti-inflammation, antitumour, and antimicrobial resource (Obied et al 2008; Rotondi et al 2010).
Many studies have confirmed the correlation between the consumption of virgin olive oil and human health promotion (reviewed in Foscolou et al 2018); however, these compounds are synthesised in a very low concentration, which in turn increases the demand for their yield improvement. Unlike other refined olive oils, the virgin olive oil refining process does not involve the use of chemicals\solvents, which would result in chemical composition variation between the refined oils and the virgin olive oils. Among the 200 phytochemicals that contribute to the conformation of the chemical profile of olive oil, triacylglycerols (unsaturated), phenolic compounds and tocopherols, are the main constituents of virgin olive oil. Nevertheless, this chemical profile is largely influenced by many factors including, cultivar, exposure to stress, and agronomical factors, and therefore, not all extracted olive oils have the same chemical profile or biological activities (Obied et al 2008; Rotondi et al 2010; Foscolou et al 2018).
Little is known about the specialised secondary metabolites biosynthesis of these three plants. For example, whilst Martin et al (2010) have identified some sesquiterpene synthase encoding genes in grape, their analysis was based on biochemical reactions, with no description of gene cluster organization underlying the identified enzymes. In recent years, many terpene synthases have been successfully characterised from plants, fungi and bacteria using whole genome sequencing (Medema and Osbourn 2016; Nützmann et al 2016; Töpfer et al 2017; Polturak and Osbourn 2021; Al-Salihi et al 2021). In this context, our study employed in silico genome mining to explore the diversity and the phylogenetic relationship of the terpene biosynthetic gene clusters across three plant genomes: grape (V. vinifera), strawberry (F. vesca), and olive (O. europaea), expanding the publicly available genomic data underlying terpene-like compounds biosynthesis within the three selected plants.
Materials and methods
Prediction of the secondary metabolite biosynthetic gene clusters
Given its efficiency in biosynthetic gene cluster prediction, the antiSMASH tool (Reddy et al 2020) was used to investigate the genomic data of three selected plants: grape (V. vinifera), strawberry (F. vesca), and olive (O. europaea) for secondary metabolite biosynthetic gene clusters, particularly terpene-like compounds. The unannotated genomes were first downloaded from either NCBI or JGI reservoirs, then submitted to the antiSMASH platform and the overall structure of all secondary metabolites biosynthetic gene clusters were predicted.
Functional annotation
To examine whether the selected genes are unique to the selected species or distributed within a small or large number of species, the predicted gene clusters of the selected species were further annotated via Blast search against the NCBI platform. The annotation was completed by the individual Blast search of the sequences of the selected enzymes against the NCBI website, using appropriate model and parameters of the standard protein BLAST suite including non-redundant protein sequences for the type of the database search, maximum 100 match for the target sequences, the expected threshold was 0.05, for the scoring parameters; matrix = BLOSUM62, gap costs = existence: 11 extension: 1, and the compositional adjustments = conditional compositional score matrix adjustment, so the most likely homologous sequences could be obtained.
Phylogenetic analysis
The MegaX software (Blin et al 2017) was used to construct the phylogenetic tree for the selected terpene synthases of the three species. The most appropriate model for the aligned sequence was estimated using the following parameters, for example, to test the tree reliability we used the bootstrap method with an average of 50 replicates, and the gaps\missing data were treated with partial deletion, while 95% was used to define the site coverage for all alignment positions. The initial tree was automatically estimated by selecting the option neighbour-Join and BioNJ and then applying the Nearest-Neighbor-Interchange parameter.
Results and discussion
Secondary metabolite profile of V. vinifera, F. vesca and O. europaea and their associated biosynthetic gene clusters
In total, 24 terpene-like genes were predicted in the three selected genomes and symbolised as G1-G10, S1-S12 and O1-O2 for grape, strawberry and olive respectfully. These preliminary findings were further verified via the Interpro platform and an extensive literature search on the mentioned compounds, revealing their chemical and biological properties as well as their pharmaceutical potentials and are described in the below sections and Table 1.
Cycloartenol
Cycloartenol is a triterpene precursor that undergoes sequential enzymatic transformation (mainly oxidative reactions), to ultimately lead to functional sterol compounds, such as ergosterol and cholesterol. Cycloartenol synthase is evolutionary related to the enzyme squalene cyclase which converts S-2,3-epoxysqualene to lanosterol in fungi (Godio et al 2004) and converts squalene to diplopterol in bacteria (Xue et al 2012; Van Der Donk 2015). The key enzymes for the chemical transformation of sterol like compounds include the sterol-4-alpha-methyl-oxidase (erg25) which is responsible for the accumulation of 4, 4-dimethyl-zymosterol. The second enzyme is 4 alpha-carboxysterol-3 beta- hydroxysteroid dehydrogenase (erg26), which belongs to the short-chain dehydrogenase/reductase family. The third enzyme is the steron reductase (erg27), which is responsible for the production of the 4-desmethyl sterol derivative. The other enzymes included in this multi-enzymatic system is the cytochrome P450. Pharmacologically, cycloartenol has shown a promising capability as antitumor, antioxidant, antimicrobial agent as well as a key player in plant growth (Dhar et al 2014; Darnet and Schaller 2019). As a phytosterol derivative, cycloartenol represents a key precursor for the biosynthesis of many sterol derivative compounds, and reports on its specific enzyme in seaweeds and algae have been published (Girard et al 2021); however, very little is known about their exact biogenetic routes that deliver their building blocks in grape specifically and in plants generally, and this is mainly because of their low yield during normal conditions. A blast search of the cycloartenol synthase sequences against the NCBI platform revealed other potential producers of this compound, amongst which Rosa chinensis, Prunus dulicis, and Quercus lobata have shown the highest sequences similarities (over 60%). These findings suggest the potential of many overlocked species as bioactive molecules producers, and can guide more experimentally verified data in planta, to better characterize our predicted metabolic pathway of such medically important compounds.
Nerolidol or 3, 7, 11-trimethyl-1, 6, 10-dodecatrien-3ol
Nerolidol is an alcoholic alkene type of sesquiterpene molecules, and very often considered as an analogue of linalool. It is naturally distributed in different types of flowers and plants. A large number of studies have confirmed the effectiveness of nerolidol as antimicrobial, antioxidant, antianxiety, anticancer as well as anti-nociceptive. In addition to its medicinal uses, it is also used as a flavouring and fragrance additive (McGinty et al 2010; Marques et al 2011; Ferreira et al 2012; Krist et al 2015). Properties like the presence of a double bond and an asymmetric centre at the C-6 and C-3 positions respectively, resulted in the conformation of four isomeric forms of nerolidol. In terms of its biosynthesis, nerolidol is naturally synthesised during the conversion of farnesyl diphosphate to the 3S-(E)-nerolidol, via nerolidol synthase that is present in the cytosol. Although nerolidol synthase was previously described in Zea mays and Celastrus angulatus (Richter et al 2016; Li et al 2021a, b), our results demonstrate the first report of this gene in two species: grape and strawberry, as well as other three potential producers of nerolidol including the species Camellia sinensis, Acer yangbiense, and Nyssa sinensis, paving the road for further derivatisation and production of many pharmaceutically useful products.
Valencene
Valencenes a rare sesquiterpene that is known for its aromatic sweetness. It is naturally occurring in citrus-scented plants and their fruits, including Valencia orange, grapefruit, nectarines, and tangerine. Valencene has many therapeutic properties, such as antiallergy, antiinflammation and antioxidant. It was also found to improve the chemotherapeutic properties of some chemotherapy drugs such as doxorubicin. Other uses of valencene include as a cleaning agent, insecticide, in cosmetics, and for food flavour. Like many terpenes, valencene is synthesised directly from the farnesyl diphosphate which is the branch point in the mevalonate pathway (Cankar et al 2015) for the synthesis of many isoprene and squalene derivatives. Of the three investigated genomes, only grape exhibited the presence of a likely associated biosynthetic gene for valencene production. On the other hand, the blast search of valencene synthase against the NCBI database has revealed the presence of more potential producers such as Populus trichocarpa, Liquidambar formosana, and Castanea mollis. Valencene yield improvement was attempted via the heterologous expression of valencene cyclase in microorganisms, mainly Saccharomyces cerevisiae (Chen et al 2019; Quyang et al. 2019). However, these studies involved no biosynthetic gene cluster characterization, and low yield of end product, indicating the need for further metabolic engineering of more associated\modifying biosynthetic gene clusters.
Farnesene
Farnesene is a sesquiterpene-like compound with the basic chemical structure C15H24, and mostly found in plant oils, especially aromatic ones such as orange and rose. In terms of its commercial uses, farnesene has been used as an enhancer element in diesel fuel, and as fragrance in cosmetics, in addition to its uses in pharma industry (Cui et al 2019; Liu et al 2020). Farnesene is one of the sesquiterpenes that requires only one enzyme to be synthesised from the ubiquitous precursor farnesyl diphosphate via the mevalonate pathway (Asadollahi et al 2008; Liu et al 2021). In our investigation, a farnesene synthase could be predicted in the grape genome, in addition to three more species including Nyssa sinensis, Pistacia vera, and Carpinus fangiana. Previous reports of the production of farnesene were mainly focused on engineering farnesene synthase in heterologous hosts, particularly in the model organisms Escherichia coli and S. cerevisiae, to increase its total yield (Nguyen et al 2012; Li et al 2018; Rahman et al 2018;); however, no biosynthetic manipulation was included in those studies.
Beta-amyrin
Beta-amyrin is a pentacyclic triterpenoids member that is rich in fatty acids, and formally known as 3beta-olean-12-en-3-ol. There are three forms of amyrin: alpha, beta, and sigma, sharing the same chemical formula C30H50O. The building blocks for beta-amyrin is oleanane molecules; concurrently, it serves as precursor for several compounds including erythrodiol, 24-hydroxy-beta-amyrin, and glycyrrhetaldehyde (Liu et al 2019). Beta-amyrin can be naturally found in some types of food including pepper, wakame and thistle. Pharmacologically, beta-amyrin is a potential anticancer, antioxidant, as well as a lipid profile enhancer (Viet et al 2021). Here, four amyrin biosynthetic core genes were predicted in both grape and strawberry genomes (two for each genome), as well as one beta-amyrin synthase in the following species, Quercus lobata, Castanea mollissima, and Malus domestica. Amyrin synthase engineering was attempted by Zhang et al (2015) to enhance the yield of amyrin in the cytosolic parts of S. cerevisiae, using the acetyl-CoA pathway, however, it was to a limited extent, due to the high cost of precursors supplementation combined with low yield improvement, making it unattractive approach for an industrial scale production of amyrin like compounds (Takemura et al 2017).
The diterpene member ent-copalyl diphosphate (ent-CPP) or copalyl diphosphoric acid
This compound also rich in fatty acids, and can be naturally found in sweet basil, eggplant, German camomile, and cardoon (Rudolf et al 2016; Zhang et al 2020). In terms of its medical uses, it has the potential of being a biomarker molecule. There are not many studies on the biological pathway of ent-CPP in plants, however, a previous bioinformatic analysis of ent-CPP sequences from Streptomyces platensis, postulated its role in converting geranyl geranyl diphosphate to ent-CPP, which in turn can be cyclised into two potent antibiotics, platensimycin and platencin (Huang et al 2014). In our bioinformatic analysis one enzyme for ent-CPP could be predicted in the grape genome, and other three unannotated genomes belong to the species Quercus lobata, Nyssa sinensis, and Juglans regia, respectively.
Germacrene
Germacrene is a sesquiterpene compound that is chemically known as (e,e)-germacra-1(10), 4,7 (11)-triene. The presence of the cyclodecane ring in germacrene helped in differentiating it from other volatile organic molecules. There are five isomers of germacrene (A-G), amongst which, germacrene A and D are the most prominent ones (Lücker et al 2004; Hu et al 2017). The medical uses of germacrene include inhibition of tumours, free radicals fighter, microbial inhibition, in addition to its uses as a food additive and in flavouring. Previous investigation of germacrene D synthase involved cloning the gene from a number of species including Lycopersicon hirsutum, Rosa hybrida, Solidago canadiensis, and Ocimum basilicum (Prosser et al 2004); however, in our research, we were able to predict four different core enzymes that are located in different loci of strawberry’s genome, and we could also predict homologous genes in the following species, Prunus dulcis, Rosa chinensis, and M. domestica. Revealing such possible pathways will probe future studies on expanding the spectrum of medicinally valuable germacrene like compounds in plants.
Alpha-pinene
Alpha-pinene is a bicyclic monoterpene-like compound which is also known as acintene. It has two forms: the alpha and beta pinene, with the chemical formula C10H16; both are among the main representatives of the volatile profile of coniferous trees. Due to their bicyclic nature, pinenes participate in the cyclization of many organic molecules including geraniol, citral, citronellal, linalool, as well as their applications as antimicrobial, anticancer, antioxidant, food additive, perfume ingredient, and as precursors for bakery and polymeric products (Vespernnann et al. 2017; Winnacker 2018; Salehi et al 2019). Amongst the three analysed genomes, only strawberry showed the presence of one biosynthetic core gene that is likely responsible for the production of pinene molecules. Additionally, three more species were predicted as potential producers of alpha-pinene; those included, Prunus dulcis, Rosa chinensis, and M. domestica. Although, there were many reports on the chemical characterisation of alpha-pinene from different plants and fungi, no genetic background could be linked to their specific biosynthetic gene clusters (Sarria et al 2014; Papa et al 2017; Niu et al 2018; Weston-Green 2021). Our in silico analysis, therefore, represents a promising start for in depth research of monoterpenes production modification and improvement in general, and alpha-pinene in particular.
Lupeol
Lupeol is a triterpene molecule with the chemical formula C30H50O that is naturally synthesized in many plants including wild carrot, canola, European plum, wild rice, green pepper, olives, strawberry, and grapes. In terms of its chemical properties, lupeol has several functional groups (e.g. methyl and hydroxyl), and one olefinic function, resulting in its pentacyclic nature. Like many other terpenoids, lupeol exhibits many biological activities, including activities against tumours, pathogenic microbes, inflammation, free radicals, heart disorders, arthritis, renal and hepatic toxicities (Saleem 2009; Li et al 2021a, b; Chóez-Guaranda et al 2021). Two enzymes that are likely associated with lupeol synthesis could be predicted in strawberry’s genome. The other three species that seems to have lupeol synthase in their genome were Rosa chinensis, Pyrus bretschneideri, M. domestica, Prunus armeniaca, Juglans regia, and Punica granatum. Due to its wide spectrum pharmacological properties, lupeol production improvement was investigated by many researchers, where different sources of the lupeol synthase, and different host organisms were attempted (Lin et al 2016; D'Adamo et al 2019; Qiao et al 2019). Again, no specific BGCs were evaluated genomically or evolutionarily.
Iridoid
Iridoid is a member of a large group of cyclopentane pyran monoterpene compounds that are composed of two carbon substitutions: secoiridoid and iridoid (Wang et al. 2020). They are widely distributed in the dicotyledonous species, mainly in Oleaceae, Labiatae, Pyrolaceae, and Rubiaceae. The biosynthesis of iridoid compounds mainly originates from the ubiquitin precursor geranyl pyrophosphate, followed by a series of reactions such as oxidation, reduction, and methylation (Leisner et al. 2017; Kouda et al. 2020). The resulting iridoid in turn will go through further modification: cyclopentane ring opening, carbonyl group oxidation and phenolic moiety conjugation, to produce the secoiridoid derivatives. This putative pathway is proposed to specifically occur in the Oleaceae family; however, no experimental evidence on their specific reactions has been reported (Bai et al. 2018). Due to the presence of the above-mentioned functional groups (mostly hydroxyl groups), iridoids can be derived into several glycosidic forms, including secoiridoid glycosides, iridoid glycosides, bis-iridoid and non-glycosidic iridoids. This structural diversity has expanded their pharmaceutical effects, especially as anti-inflammation, anticancer, antioxidant agents (Cádiz-Gurrea et al. 2021). Our gene function annotation revealed the presence of one predicted iridoid synthase in olive’s genome among the three investigated genomes, as well as three other species: Handroanthus impetiginosus, Phtheirospermum japonica, and Buddleja alternifolia.
A brief description of the functional properties of the putative enzymes which were predicted using the Interpro database, including description of enzyme function, sequence identity, and their homologous enzymes in other species, is listed in Table 1.
Biosynthetic gene clusters prediction
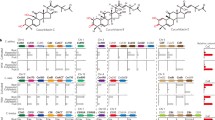
To investigate whether grape, strawberry and olive, have biosynthetic gene clusters in common, the genomes of the three species were mined, and several secondary metabolite biosynthetic gene clusters were predicted. An overall of the predicted biosynthetic gene clusters is presented in Table 2 and Figs. 1, 2 and 3. Of the 46 predicted clusters in V. vinifera (grape), 10 appear to be associated with terpene-like compounds production, including cycloartenol, nerolidol, valencene, alpha-farnesene, beta-amyrin, copalyl diphosphate, oxidosequalene, isoprene, ent-kaur-16-ene, while of the 33 predicted clusters in F. vesca (strawberry), 12 were related to terpenoid like compounds, including ent-kaur-16-ene, germacrene, beta-amyrin, alpha-pinene, lupeol, franesene and nerolidol. Finally, of the 14 predicted biosynthetic gene clusters in O. europaea (olive), two were most likely correlated to terpene derivatives such as alpha-farnesene and iridoid.
Grape biosynthetic gene clusters annotation
Sequences analysis of the grape genome resulted in the prediction of ten putative terpene BGCs that are illustrated in the following sections and Fig. 1.
-
1.
Two cycloartenol\oxdiosequalene BGCs (A & B) were predicted in the grape genome. Cluster A spans 73 kb, and consists of five genes, four of which were predicted as cycloartenol synthase, and one as hypothetical or functionally uncharacterised protein. The presence of several cycloartenol synthase suggest that some of these genes might be duplicated or pseudogenes, or they might be misassembled during the genome assembly process. On the other hand, the other cycloartenol BGC (cluster B), was predicted to have eight biosynthetic genes within its 330 kb DNA region. Biosynthetic genes like serine carboxypeptidase and cytochrome P450 were among the predicted genes in this cluster. Whole cluster blast search via antiSMASH platform for cluster A revealed no significant similarity with other BGCs in other species; however, for cluster B, we could observe one homologous BGC that is located in the genome of Gossypium raimondii on chromosome 9, suggesting familiar synthesis path for cycloartenol and its derivatives in this species.
-
2.
Nerolidol BGC included six genes, however, all of them were predicted as nerolidol synthase. The cluster was predicted to occupy a 70 kb region of the genome. The blast search against other species genomes, returned with almost a homologous (60% similarity) BGC that is located on the chromosome-16 of the Manihot esculenta genome.
-
3.
In the valencene BGC, seventeen biosynthetic genes were predicted, however, and similarly to the nerolidol BGC all were predicted as valencene synthase. The borders of this cluster were overlapping a 737 kb region. Several BGCs of unannotated genomes of other species appeared to share high sequence similarity with this predicted BGC, including 100% with Arachis ipaensis on chromosome 8, 71% with Citrus sinensis on chromosome 4 and 74% with Eucalyptus grandis extracted from phytozome platform.
-
4.
Alpha-farnesene is of a 43 kb size BGC that was predicted to involve eight biosynthetic gene clusters, of which two were predicted as alpha-farnesene synthase, and two as anthocyanidin reductase, the other four were functionally unknown and annotated as hypothetical protein. Genome comparison with other species, revealed the presence of two homologous BGCs, one was located in Theobroma cacao genome on chromosome 7 and the other one was located in Glycine max genome on chromosome 82; both had 50% sequence similarity with our annotated cluster.
-
5.
Two putative beta-amyrin BGCs (A & B) could be predicted in the investigated grape genome. Cluster A consisted of nine biosynthetic genes and occupied 46 kb region of the DNA. In addition to the core enzyme amyrin synthase, a beta-amyrin oxidase was predicted. The remaining genes were annotated as hypothetical proteins. Two separated BGCs, one on chromosome 4 in Aquilegia coerulea, and one on chromosome 10 in Raimondii chromosom, have shown 70% and 60% sequence similarity respectively with our beta-amyrin BGC-A. While, cluster B organisation, involved fifteen biosynthetic genes, that spans 391 kb. In addition to the beta-amyrin synthase, a gibberellin beta dioxygenase was also predicted within this cluster as well as several functionally unknown genes. Apparently, amyrin derivatives production is common in plants as our cluster analysis has revealed many and almost identical BGCs that are located in genomes of different species of which, T. cacao chromosome 6, A. coerulea chromosome 4, and P. persica, have shown 100% sequence similarity, and Gossypium raimondii chromosome 9, Solanum lycopersicum chromosome 12, Camelina sativa chromosome 9, Arabidopsis lyrata, Capsella grandiflora extracted from phtozome, Boechera stricta extracted from phytozome, have shown 60% sequence similarity.
-
6.
Copalyl diphosphate BGC, had a size of 93 kb, in which eight biosynthetic genes were annotated. Those genes were predicted as copalyl diphosphate synthase, N-terminal acetyltransferase, and hypothetical proteins. This predicted BGC has shown 66% sequence homologous with both Prunus persica and G. raimondii on chromosome 4, as well as 50% sequence homologous with the Populus trichocarpa chromosome 2.
-
7.
Isoprene BGC, ten biosynthetic genes were annotated in this 211 kb cluster, including geranyltransferase and several hypothetical proteins, similarity to the cycloartenol BGC, isoprene BGC displayed a 50% sequence similarity with a BGC from the Citrus sinensis on chromosome 2.
-
8.
Ent-kaurene BGC is a hybrid cluster of terpene and saccharide compounds biosynthesis, with the size of 150 kb, that is include nine genes. Anthocyanidin 3-O-glucosyltransferase and 2-dehydro-3-deoxy phosphooctonate aldolase were among the annotated key enzymes in this cluster. Ent-kaurene BGC showed the following sequence similarity 62%, 50%, 50% and 50% with other BGCs located on genomes of the species, Arabidopsis thaliana chromosome 1, Camelian sativa chromosome 16, Boechera stricta scaffold 20129 extracted from phytozome, Populua trichocarpa chromosome 8, respectively.
Strawberry gene clusters annotation
For the strawberry genome analysis, twelve terpenoid BGCs were predicted and described in the below sections and Fig. 2.
-
1.
Two separate Alpha-pinene BGCs (A&B) were predicted in the strawberry genome. For cluster A, eleven biosynthetic genes could be annotated and occupied in a 36 kb length. The closest adjacent gene to the alpha-pinene synthase was predicted as aminocyclopropane-1-carboxylate oxidase as well as several hypothetical proteins. No homologous BGC could be found in other species during our sequence blast analysis. For alpha pinene BGC-A, it was predicted to be 155 kb involving twelve biosynthetic genes. Functional annotation of its genes uncovered the presence of several modifying genes, such as oxidoreductase and aminocyclopropane-1-carboxylate oxidase. Similarly to cluster A, no significant sequence similarity in other species could be found in the database for cluster B.
-
2.
Ent-kaurene BGC identified as a cluster of nine biosynthetic genes, with border length of 96 kb. The cluster consisted of three ent-kaurene synthases as well as one cytochrome P450 and a number of hypothetical proteins. Cluster sequences blast against other species, has returned with no significant similarities.
-
3.
Germacrene D, four biosynthetic gene clusters A, B, C, and D with the following sizes, 116 kb, 127 kb, 114 kb, and 37 kb, appeared to have the germacrene-D synthase as its core enzyme, among which only one BGC showed 50% sequence similarity with a BGC on chromosome 5 of the Citrus sinensis genome. Several modifying genes such as methyltransferase, carboxylesterase, acyltransferase, cytochrome P450 and aminocyclpropane-1-carboxylate oxidase were also predicted in these clusters.
-
4.
Beta amyrin BGC annotation revealed the size of this cluster as 171 kb, and the presence of five biosynthetic genes including the beta amyrin synthase. Two of these biosynthetic genes were predicted as cytochrome P450s, and the other two were predicted as hypothetical proteins. Sequences similarity investigation revealed the presence of many homologous biosynthetic genes in different species with the following percentages, 71% in each of Camelina sativa on chromosome 9, Capsella grandiflora scaffold 1361, Boechera stricta scaffold 20129, and 57% in both Brassica rapa and Arabidopsis lyrate extracted from phytozome, and 85% and 100% in Phaseolus vulgaris on chromosome 10, and P. persica respectively.
-
5.
Nerolidol BGC is a biosynthetic cluster that consists of eleven genes 65 kb in length. Anthocyanidin reductase was one of the predicted biosynthetic genes as well as several hypothetical proteins. A putative BGC with 57% homologous genes was found in P. persica genome.
-
6.
Two Lupeol\oxidosequalene BGCs (A&B) with the length of 73 kb and 150 kb respectively, were predicted. There clusters structure involved nineteen and seven biosynthetic genes, in addition to their core enzyme. Those genes included NADH ubiquinone oxidoreductase, shikimate-O-hydroxycinnamoyltransferase, 2- oxoglutarate dependent dioxygenase, and a number of functionally unknown genes.
-
7.
Tricyclene beta ocimene BGC length was predicted to be 52 kb which included six genes within its border. Cytochrome P450 and coumaroyltranferase genes, were among those predicted genes.
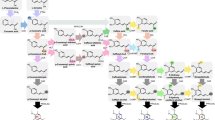
Olive gene clusters annotation
The olive genome appeared to have two terpene BGCs, as described below and in Fig. 3.
-
1.
Iridoid BGC was predicted to span a DNA region of 249 kb, and consist of 12 candidate genes; cytochrome P450 and lipoxygenase were among the predicted modifying genes. No significant sequence similarities could be detected in our blast search against other species genomes.
-
2.
Alpha farnesene BGC, in this 180 kb BGC a total of 13 biosynthetic genes were predicted. Several tailoring genes were putatively annotated including cytochrome P450, and Gibberellin-2- beta-dioxygenase. Similarly, to iridoid BGC, no homologous BGC could be predicted during the sequences analysis.
Phylogenetic tree analysis
The MegaX software was used to construct the phylogenetic tree for the selected terpene synthases of the three species. The most appropriate model for the aligned sequence was estimated using the following parameters, for example, we used the bootstrap method with an average of 50 replicates to test the tree reliability, and the gaps\missing data were treated with partial deletion, while 95% was used to define the site coverage for all alignment positions. The initial tree was automatically estimated by selecting the option neighbour-Join and BioNJ and then applying the Nearest-Neighbor-Interchange parameter. Our tree was constructed using the sequences of the predicted terpene synthase of the three species (total = 24 genes), to investigate the potential of sequence homology among the selected species.
It is known that taxonomically related plants produce chemically similar yet structurally divergent specialised secondary metabolites, and often, this structural diversity can alter the pharmaceutical and the biological properties of these organic molecules. These chemical and biological differences in plants’ end products, might be due to their adaptation response to their environment’s selective pressure, such as pathogens and insect attacks, as well as random genetic drift (Frey et al 1997; Fan et al 2020). Therefore, more future BGCs related investigations will provide closer insight into the mechanisms and pathways that those secondary metabolites utilise during the interaction between their native producers and the surrounding environment and could propose gene sets for the improvement of crop and medicinally important compounds production through metabolic engineering in heterologous organisms such as tobacco, yeast, and E. coli (Lin et al 2016; Calegario et al 2016; Owen et al 2017).
Five terpene synthases including germacrene D (S2), germacrene D (S3), germacrene D (S4), germacrene D (S5), alpha pinene (S7) from strawberry were clustered in clade 1 with valencene (G3) from grape as a close relative, indicating their similarity in terms of their catalytic activity. As for clade 2, it consisted of five terpene synthases; two of them nerolidol (S12) and alpha-farnesen synthase (S9) were from strawberry, and the other two Isoprene (G9) and alpha-farnesene (G4) from grape which was clustered closely with terpene synthase (O2) from olive, and shared 100% sequence similarity with it, suggesting their identical cyclization pattern. In contrast to clade 1, clade 3 included three terpene synthases from grape nerolidol (G2), copalyl diphosphate (G6), ent-kaurene (G10) and one enzyme ent-kaurene (S1) from strawberry, of which G10 and S1 were 100% identical. The remaining enzymes for both strawberry beta-amyrin (S8), copalyl diphosphate 6, lupeol 11, lupeol 10 and grape G5 beta amyrin, amyrin synthase7, cycloartenol 8, cycloartenol 1 were placed together in clade 4, again, proposing their cyclization types of similarity and their evolutionary relationship (see Fig. 4 for more details). It can be said that our tree demonstrates the evolutionary relationship between the compounds described above, as despite the three selected species being taxonomically unrelated, many enzymes that putatively produce structurally diverse chemicals were clustered in one clade, indicating that these genes are evolutionarily conserved and that they have similar motifs. The prediction of such sequences and mode of action similarities motivates future metabolic engineering of the selected enzymes and their end products.
Maximum likelihood tree of the core terpene enzymes of the three species, grape, strawberry, and olive. Grape terpene synthases labelled as = Cycloartenol-A (G1), Nerolidol (G2), Valencene (G3), Alpha-farnesene (G4), Beta-amyrin (G5), Copalyl diphosphate (G6), Beta-amyrin synthase (G7), Cycloartenol-B (G8), Isoprene (G9), Ent-kaurene (G10). Strawberry terpene synthases labelled as= alpha-pinene (S1), Ent-kaurene (S2), Alpha pinene (S3), Germacrene-D1 (S4), Germacrene-D2 (S5), Germacrene-D3 (S6), Germacrene-D4 (S7), Alpha-pinene (S8), Beta amyrin (S9), Beta-ocimene (S10), Lupeol (S11), Nerolidol (S12). Olive terpene synthases labelled as = Iridoid (O1), Alpha-farnesene (O2)
Conclusion
Plants represent inexhaustible source of specialised secondary metabolites. These secondary metabolites can provide protection for their producers against microbial pathogens or herbivores. In addition to their ecological roles secondary metabolites can serve as drug precursors in the drug industry. The first BGC in plants was reported two decades ago (Frey et al 1997; Kumar et al 2016a, b), ever since, many clusters have been described, as more sequencing tools developed and become accessible. The chemical profile of the three species has been broadly studied, and a wide range of medicinally valuable compounds have been described, however, very little is known about their genetic background. To this end a BLASTp search was carried out using the sequence of the core enzymes that have been identified via antiSMASH tool, to possibly identify the exact compounds that are likely produced by those enzymes. This analysis has enabled us to highlight a group of core enzymes that are correlated to the production of molecules including cycloartenol, nerolidol, valencene, farnesene, beta-amyrin, ent-copalyl diphosphate, germacrene, alpha-pinene, lupeol and iridoid.
The three species selected here produce monoterpene and triterpenes derived compounds, and in our genome mining, we observed subtle, but possibly important pathway derivatisation, that could possibly lead to structural modification of their end products. By annotating and comparing the key enzymes and their BGCs with other species, we could putatively correlate the genomic structure of many previously overlocked pharmaceutically valuable compounds, such as cycloartenol, nerolidol, valencene, farnesene, beta-amyrin lupeol in three globally important species. Such in-silico analysis offers compelling evidence of genomic features of many medically important molecules, and putatively linking them to their specific biosynthetic gene clusters and substituting many laborious in vitro screening of certain enzymes. Moreover, our gene function annotation, and comparative biosynthetic gene cluster analyses shed light on the genetic background and the putative biochemical functions of many previously uncharacterised pathways in the selected plants as well as other species.
Despite the abundant chemical profiles of our selected plants, their biosynthesis pathways remain unsolved. Our putative BGCs have been characterised for many compound scaffolds, especially pathways of tri-di-sesquiterpenoids biosynthesis. The abundance of these BGCs was diverse across the selected species, as they ranged from 2 to 10 or more, such clusters may occupy a genomic regions of 36 kb up to several hundreds of kilobytes to include five to 20 candidate genes within their borders. Most of our BGCs encoded enzymes that are specialised in catalysing the production of unique backbones for the synthesis of specific specialised metabolites, such as the rare valencene and the multifunctional cycloartenol. In addition to the core enzymes, our BGCs collections included tailoring genes and regulatory elements (e.g., cytochrome P450, methyltransferase and several oxido-reductase genes) to further modify the synthesised backbones and channel them into specific pathways. The elucidation of these BGCs therefore, will facilitate the investigation\discovery of several sophisticated metabolic pathways and further expand their chemical diversification\properties, as well as the biogenetic origin of many related plants secondary metabolites. It can also define the role of those secondary metabolites in the interaction between the plants and their competitive inhabitants (e.g., insects and microbes), and ultimately provide solutions for enhancing their adverse biotic\abiotic conditions. Our investigation represents a step forward towards the characterisation\modification of potential novel chemicals, that are of urgent need to defeat the arising antibiotic resistance.
Abbreviations
- BCG:
-
Biosynthetic gene cluster
- ent-CPP:
-
Ent-copalyl diphosphate
References
Agrawal AA, Heil M (2012) Synthesizing specificity: multiple approaches to understanding the attack and defense of plants. Trends Plant Sci 17(5):239–242. https://doi.org/10.1016/j.tplants.2012.03.011
Al-Salihi SAA, Alberti F (2021) Naturally occurring terpenes: a promising class of organic molecules to address influenza pandemics. Nat Prod Bioprospect 11:405–419. https://doi.org/10.1007/s13659-021-00306-z
Al-Salihi SAA, Bull ID, Al-Salhi R, Gates PJ, Salih KS, Bailey AM, Foster GD (2021) Further biochemical profiling of Hypholoma fasciculare metabolome reveals its chemogenetic diversity. Front Bioeng Biotechnol. https://doi.org/10.1101/2020.05.28.122176
Al-Salihi SAA, Dao TT, Williams K, Bailey AM, Foster GD (2019) The biogenetic origin of the biologically active naematolin of Hypholoma species involves an unusual sesquiterpene synthase. Mol Biotechnol 61(10):754–762. https://doi.org/10.1007/s12033-019-00199-x
Asadollahi MA, Maury J, Møller K, Nielsen KF, Schalk M, Clark A, Nielsen J (2008) Production of plant sesquiterpenes in Saccharomyces cerevisiae: effect of ERG9 repression on sesquiterpene biosynthesis. Biotechnol Bioeng 99(3):666–677. https://doi.org/10.1002/bit.21581
Bai M, Li SF, Liu SF, Wang XB, Huang XX, Song SJ (2018) Iridoid glycoside and lignans from a wild vegetable (Patrinia villosa Juss.) with antioxidant activity. J Food Biochem 42(3):e12521. https://doi.org/10.1111/jfbc.12521
Banerjee A, Hamberger B (2018) P450s controlling metabolic bifurcations in plant terpene specialized metabolism. Phytochem Rev 17(1):81–111. https://doi.org/10.1007/s11101-017-9530-4
Bathe U, Tissier A (2019) Cytochrome P450 enzymes: a driving force of plant diterpene diversity. Phytochem 161:149–162. https://doi.org/10.1016/j.phytochem.2018.12.003
Black CA, Parker M, Siebert TE, Capone DL, Francis IL (2015) Terpenoids and their role in wine flavour: recent advances. Aust J Grape Wine Res 21:582–600. https://doi.org/10.1111/ajgw.12186
Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, De Los Santos EL, Kim HU, Nave M, Dickschat JS (2017) antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45(1):36–41. https://doi.org/10.1093/nar/gkx319
Bohlmann J, Keeling CI (2008) Terpenoid biomaterials. Plant J 54(4):656–669. https://doi.org/10.1111/j.1365-313X.2008.03449.x
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Booth JK, Bohlmann J (2019) Terpenes in Cannabis sativa–from plant genome to humans. Plant Sci J 284:67–72. https://doi.org/10.1016/j.plantsci.2019.03.022
Cádiz-Gurrea MDLL, Pinto D, Delerue-Matos C, Rodrigues F (2021) Olive fruit and leaf wastes as bioactive ingredients for cosmetics—a preliminary study. Antioxidants 10(2):245. https://doi.org/10.3390/antiox10020245
Calegario G, Pollier J, Arendt P, de Oliveira LS, Thompson C, Soares AR, Pereira RC, Goossens A, Thompson FL (2016) Cloning and functional characterization of cycloartenol synthase from the red seaweed Laurencia dendroidea. PLoS One 11(11):1e0165954. https://doi.org/10.1371/journal.pone.0165954
Cankar K, Jongedijk E, Klompmaker M, Majdic T, Mumm R, Bouwmeester H, Bosch D, Beekwilder J (2015) (+)-Valencene production in Nicotiana benthamiana is increased by down-regulation of competing pathways. Biotechnol J 10(1):180–189. https://doi.org/10.1002/biot.201400288
Chen H, Zhu C, Zhu M, Xiong J, Ma H, Zhuo M, Li S (2019) High production of valencene in Saccharomyces cerevisiae through metabolic engineering. Microb Cell Fact 18(1):1–14. https://doi.org/10.1186/s12934-019-1246-2
Chóez-Guaranda I, García J, Sánchez C, Pesantes C, Flores J, Manzano P (2021) Identification of lupeol produced by Vernonanthura patens (Kunth) H. Rob. leaf callus culture. Nat Prod Res 35(3):503–507. https://doi.org/10.1080/14786419.2019.1636239
Christianson DW (2008) Unearthing the roots of the terpenome. Curr Opin Chem Biol 12(2):141–150. https://doi.org/10.1016/j.cbpa.2007.12.008
Cui Z, Jiang X, Zheng H, Qi Q, Hou J (2019) Homology-independent genome integration enables rapid library construction for enzyme expression and pathway optimization in Yarrowia lipolytica. Biotechnol Bioengin 116(2):354–363. https://doi.org/10.1002/bit.26863
D’Adamo S, Schiano di Visconte G, Lowe G, Szaub-Newton J, Beacham T, Landels A, Allen MJ, Spicer A, Matthijs M (2019) Engineering the unicellular alga Phaeodactylum tricornutum for high-value plant triterpenoid production. Plant Biotechnol J 17(1):75–87. https://doi.org/10.1111/pbi.12948
Darnet S, Schaller H (2019) Metabolism and biological activities of 4-methyl-sterols. Mole 24(3):451. https://doi.org/10.3390/molecules24030451
Davis EM, Croteau R (2000) Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. Biosynthesis. https://doi.org/10.1007/3-540-48146-X_2
Dellas N, Thomas ST, Manning G, Noel JP (2013) Discovery of a metabolic alternative to the classical mevalonate pathway. eLife 2:e00672. https://doi.org/10.7554/eLife.00672
Dhar N, Rana S, Razdan S, Bhat WW, Hussain A, Dhar RS, Vaishnavi S, Hamid A, Vishwakarma R, Lattoo SK (2014) Cloning and functional characterization of three branch point oxidosqualene cyclases from Withania somnifera (L.) dunal. J Biol Chem 289(24):17249–17267. https://doi.org/10.1074/jbc.M114.571919
Dunlevy JD, Dennis EG, Soole KL, Perkins MV, Davies C, Boss PK (2013) A methyltransferase essential for the methoxypyrazine-derived flavour of wine. Plant J 75(4):606–617. https://doi.org/10.1111/tpj.12224
Edger PP, Poorten TJ, VanBuren R, Hardigan MA, Colle M, McKain MR, Smith RD, Teresi SJ, Nelson AD, Wai CM, Alger EI (2019) Origin and evolution of the octoploid strawberry genome. Nat Genet 51(3):541–547. https://doi.org/10.1038/s41588-019-0356-4
Fan P, Wang P, Lou YR, Leong BJ, Moore BM, Schenck CA, Combs R, Cao P, Brandizzi F, Shiu SH, Last RL (2020) Evolution of a plant gene cluster in Solanaceae and emergence of metabolic diversity. eLife 9:56717. https://doi.org/10.7554/eLife.56717.sa2
FAO (2021) Statistical yearbook: world food and agriculture
Ferreira FM, Palmeira CM, Oliveira MM, Santos D, Simões AM, Rocha SM, Coimbra MA, Peixoto F (2012) Nerolidol effects on mitochondrial and cellular energetics. Toxicol In Vitro 26(2):189–196. https://doi.org/10.3390/molecules21050529
Foscolou A, Critselis E, Panagiotakos D (2018) Olive oil consumption and human health: a narrative review. Maturitas 118:60–66. https://doi.org/10.1016/j.maturitas.2018.10.013
Frey M, Chomet P, Glawischnig E, Stettner C, Grun S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, Simcox K (1997) Analysis of a chemical plant defense mechanism in grasses. Science 277(5326):696–699. https://doi.org/10.1126/science.277.5326.696
Girard J, Lanneau G, Delage L, Leroux C, Belcour A, Got J, Collén J, Boyen C, Siegel A, Dittami SM, Leblanc C (2021) Semi-quantitative targeted gas chromatography-mass spectrometry profiling supports a late side-chain reductase cycloartenol-to-cholesterol biosynthesis pathway in brown algae. Front Plant Sci. https://doi.org/10.3389/fpls.2021.648426
Godio RP, Fouces R, Gudina EJ, Martin JF (2004) Agrobacterium tumefaciens-mediated transformation of the antitumor clavaric acid-producing basidiomycete Hypholoma sublateritium. Curr Genet 46(5):287–294
González-Domínguez R, Sayago A, Akhatou I, Fernández-Recamales Á (2020) Multi-chemical profiling of strawberry as a traceability tool to investigate the effect of cultivar and cultivation conditions. Foods 9(1):96. https://doi.org/10.1007/s00294-004-0533-5
Hu Y, Zhou YJ, Bao J, Huang L, Nielsen J, Krivoruchko A (2017) Metabolic engineering of Saccharomyces cerevisiae for production of germacrene A, a precursor of beta-elemene. J Ind Microbiol Biotechnol 44(7):1065–1072. https://doi.org/10.3390/fermentation8090429
Huang T, Yang D, Rudolf JD, Xie P, Xie G, Teng Q, Lohman JR, Zhu X, Huang Y, Zhao LX, Jiang Y (2014) Strain prioritization for natural product discovery by a high-throughput real-time PCR method. J Nat Prod 77(10):2296–2303. https://doi.org/10.1021/np5006168
Ilc T, Werck-Reichhart D, Navrot N (2016) Meta-analysis of the core aroma components of grape and wine aroma. Front Plant Sci 7:1472. https://doi.org/10.3389/fpls.2016.01472
Jabir MS, Taha AA, Sahib UI (2018) Linalool loaded on glutathione-modified gold nanoparticles: a drug delivery system for a successful antimicrobial therapy. Artif Cells Nanomed Biotechnol 46(sup2):345–355. https://doi.org/10.1080/21691401.2018.1457535
Jia Q, Chen X, Köllner TG, Rinkel J, Fu J, Labbé J, Xiong W, Dickschat JS, Gershenzon J, Chen F (2019) Terpene synthase genes originated from bacteria through horizontal gene transfer contribute to terpenoid diversity in fungi. Sci Rep 9(1):1–11. https://doi.org/10.1038/s41598-019-45532-1
Jin D, Dai K, Xie Z, Chen J (2020) Secondary metabolites profiled in cannabis inflorescences, leaves, stem barks, and roots for medicinal purposes. Sci Rep 10(1):1–14. https://doi.org/10.1038/s41598-020-60172-6
Kalua CM, Boss PK (2010) Comparison of major volatile compounds from Riesling and Cabernet Sauvignon grapes (Vitis vinifera L.) from fruitset to harvest. Aust J Grape Wine Res 16(2):337–348. https://doi.org/10.1111/j.1755-0238.2010.00096.x
Karunanithi PS, Zerbe P (2019) Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front Plant Sci 10:1166. https://doi.org/10.3389/fpls.2019.01166
Kouda R, Yakushiji F (2020) Recent advances in iridoid chemistry: biosynthesis and chemical synthesis. Chem Asian J 15(22):3771–3783. https://doi.org/10.1002/asia.202001034
Krist S, Banovac D, Tabanca N, Wedge DE, Gochev VK, Wanner J, Schmidt E, Jirovetz L (2015) Antimicrobial activity of nerolidol and its derivatives against airborne microbes and further biological activities. Nat Prod Commun. https://doi.org/10.1177/1934578X1501000133
Kumar S, Kempinski C, Zhuang X, Norris A, Mafu S, Zi J, Bell SA, Nybo SE, Kinison SE, Jiang Z, Goklany S (2016) Molecular diversity of terpene synthases in the liverwort Marchantia polymorpha. Plant Cell 28(10):2632–2650. https://doi.org/10.1105/tpc.16.00062
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Leisner CP, Kamileen MO, Conway ME, O’Connor SE, Buell CR (2017) Differential iridoid production as revealed by a diversity panel of 84 cultivated and wild blueberry species. PLoS One 12(6):e0179417. https://doi.org/10.1371/journal.pone.0179417
Li D, Pan C, Lu J, Zaman W, Zhao H, Zhang J, Lü S (2021) Lupeol accumulation correlates with auxin in the epidermis of castor. Molecules 26(10):2978. https://doi.org/10.3390/molecules26102978
Li Q, Huang B, He Q, Lu J, Li X, Li Z, Wu H, Ye Q (2018) Production of succinate from simply purified crude glycerol by engineered Escherichia coli using two-stage fermentation. Bioresour Bioprocess 5(1):1–10. https://doi.org/10.1186/s40643-018-0227-3
Li W, Yan X, Zhang Y, Liang D, Caiyin Q, Qiao J (2021) Characterization of trans-nerolidol synthase from Celastrus angulatus Maxim and production of trans-nerolidol in engineered Saccharomyces cerevisiae. J Agric Food Chem 69(7):2236–2244. https://doi.org/10.1021/acs.jafc.0c06084
Lin J, Massonnet M, Cantu D (2019) The genetic basis of grape and wine aroma. Horti Res. https://doi.org/10.1038/s41438-019-0163-1
Lin TT, Wang D, Dai ZB, Zhang XL, Huang LQ (2016) Construction of cell factories for production of lupeol in Saccharomyces cerevisiae. Chin J Chin Mat Med 41(6):1008–1015. https://doi.org/10.4268/cjcmm20160606
Liu H, Fan J, Wang C, Li C, Zhou X (2019) Enhanced β-amyrin synthesis in Saccharomyces cerevisiae by coupling an optimal acetyl-CoA supply pathway. J Agri Food Chem 67(13):3723–3732. https://doi.org/10.1021/acs.jafc.9b00653
Liu SC, Liu Z, Wei LJ, Hua Q (2020) Pathway engineering and medium optimization for α-farnesene biosynthesis in oleaginous yeast Yarrowia lipolytica. J Biotechnol 319:74–81. https://doi.org/10.1016/j.jbiotec.2020.06.005
Liu Y, Wang Z, Cui Z, Qi Q, Hou J (2021) α-Farnesene production from lipid by engineered Yarrowia lipolytica. Bioresour Bioprocess 8(1):1–12. https://doi.org/10.1186/s40643-021-00431-0
Lücker J, Bowen P, Bohlmann J (2004) Vitis vinifera terpenoid cyclases: functional identification of two sesquiterpene synthase cDNAs encoding (+)-valencene synthase and (−)-germacrene D synthase and expression of mono-and sesquiterpene synthases in grapevine flowers and berries. Phytochemistry 65(19):2649–2659. https://doi.org/10.1016/j.phytochem.2004.08.017
Marques AM, Barreto ALS, Curvelo JADR, Romanos MTV, Soares RMDA, Kaplan MAC (2011) Antileishmanial activity of nerolidol-rich essential oil from Piper claussenianum. Revista Brasileira de Farmacognosia 21(5):908–914. https://doi.org/10.1590/S0102-695X2011005000157
Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol 10(1):1–22. https://doi.org/10.1186/1471-2229-10-226
Matarese F, Cuzzola A, Scalabrelli G, D’Onofrio C (2014) Expression of terpene synthase genes associated with the formation of volatiles in different organs of Vitis vinifera. Phytochemistry 105:12–24. https://doi.org/10.1016/j.phytochem.2014.06.007
McGinty D, Letizia CS, Api AM (2010) Addendum to fragrance material review on Nerolidol (isomer unspecified). Food Chem Toxicol 48:S43–S45. https://doi.org/10.1016/j.fct.2009.11.008
Medema MH, Osbourn A (2016) Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways. Nat Prod Rep 33(8):951–962. https://doi.org/10.1039/c6np00035e
Nassiri-Asl M, Hosseinzadeh H (2009) Review of the pharmacological effects of Vitis vinifera (Grape) and its bioactive compounds. Phyto Res 23(9):1197–1204. https://doi.org/10.1002/ptr.2761
Nguyen TD, MacNevin G, Ro DK (2012) De novo synthesis of high-value plant sesquiterpenoids in yeast. In: Abelson J, Simon M, Verdine G, Pyle A (eds) Methods in enzymology, vol 517. Academic Press, London, pp 261–278
Niu FX, He X, Wu YQ, Liu JZ (2018) Enhancing production of pinene in Escherichia coli by using a combination of tolerance, evolution, and modular co-culture engineering. Front Microb 9:1623. https://doi.org/10.3389/fmicb.2018.01623
Nützmann HW, Huang A, Osbourn A (2016) Plant metabolic clusters–from genetics to genomics. N phytologist 211(3):771–789. https://doi.org/10.1111/nph.13981
Obied HK, Prenzler PD, Ryan D, Servili M, Taticchi A, Esposto S, Robards K (2008) Biosynthesis and biotransformations of phenol-conjugated oleosidic secoiridoids from Olea europaea L. Nat Prod Rep 25(6):1167–1179. https://doi.org/10.1039/B719736E
Oldfield E, Lin FY (2012) Terpene biosynthesis: modularity rules. Angew Chem Int Ed 51(5):1124–1137. https://doi.org/10.1002/anie.201103110
Ouyang X, Cha Y, Li W, Zhu C, Zhu M, Li S, Zhuo M, Huang S, Li J (2019) Stepwise engineering of Saccharomyces cerevisiae to produce (+)-valencene and its related sesquiterpenes. RSC Adv 9(52):30171–30181. https://doi.org/10.1039/C9RA05558D
Owen C, Patron NJ, Huang A, Osbourn A (2017) Harnessing plant metabolic diversity. Curr Opin Chem Biol 40:24–30. https://doi.org/10.1016/j.cbpa.2017.04.015
Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496(7446):528–532. https://doi.org/10.1038/nature12051
Papa G, Kirby J, Konda NM, Tran K, Singh S, Keasling JD, Peter GF, Simmons BA (2017) Development of an integrated approach for α-pinene recovery and sugar production from loblolly pine using ionic liquids. Green Chem 19(4):1117–1127. https://doi.org/10.1039/C6GC02637K
Parker M, Capone DL, Francis IL, Herderich MJ (2017) Aroma precursors in grapes and wine: flavor release during wine production and consumption. J Agri Food Chem 66(10):2281–2286. https://doi.org/10.1021/acs.jafc.6b05255
Pateraki I, Heskes AM, Hamberger B (2015) Cytochromes P450 for terpene functionalisation and metabolic engineering. Biotechnol Isoprenoids. https://doi.org/10.1007/10_2014_301
Peretto G, Du WX, Avena-Bustillos RJ, Sarreal SBL, Hua SST, Sambo P, McHugh TH (2014) Increasing strawberry shelf-life with carvacrol and methyl cinnamate antimicrobial vapors released from edible films. Post Biol Technol 89:11–18. https://doi.org/10.1016/j.postharvbio.2013.11.003
Perkins-Veazie P and Huber DJ (1988). Growth and ripening of strawberry fruit under field conditions. In: Proceedings of the annual meeting of the Florida State Horticulture Society (USA)
Polturak G, Osbourn A (2021) The emerging role of biosynthetic gene clusters in plant defense and plant interactions. PLoS Pathogens 17(7):e1009698. https://doi.org/10.1371/journal.ppat.1009698
Potter D, Luby JJ, Harrison RE (2000) Phylogenetic relationships among species of Fragaria (Rosaceae) inferred from non-coding nuclear and chloroplast DNA sequences. Syst Bot 25(2):337–348. https://doi.org/10.2307/2666646
Prosser I, Altug IG, Phillips AL, König WA, Bouwmeester HJ, Beale MH (2004) Enantiospecific (+)-and (−)-germacrene D synthases, cloned from goldenrod, reveal a functionally active variant of the universal isoprenoid-biosynthesis aspartate-rich motif. Arch Biochem 432(2):136–144. https://doi.org/10.1016/j.abb.2004.06.030
Qiao W, Zhou Z, Liang Q, Mosongo I, Li C, Zhang Y (2019) Improving lupeol production in yeast by recruiting pathway genes from different organisms. Sci Rep 9(1):1–8. https://doi.org/10.1038/s41598-019-39497-4
Rahman M, Xu CC, Qin W (2018) Exotic glycerol dehydrogenase expressing Escherichia coli increases yield of 2, 3-butanediol. BIOB 5(1):1–11. https://doi.org/10.1186/s40643-018-0189-5
Reddy GK, Leferink NG, Umemura M, Ahmed ST, Breitling R, Scrutton NS, Takano E (2020) Exploring novel bacterial terpene synthases. PloS one 15(4):e0232220. https://doi.org/10.1371/journal.pone.0232220
Richter A, Schaff C, Zhang Z, Lipka AE, Tian F, Köllner TG, Schnee C, Preiß S, Irmisch S, Jander G, Boland W (2016) Characterization of biosynthetic pathways for the production of the volatile homoterpenes DMNT and TMTT in Zea mays. Plant Cell 28(10):2651–2665. https://doi.org/10.1105/tpc.15.00919
Rotondi A, Alfei B, Magli M, Pannelli G (2010) Influence of genetic matrix and crop year on chemical and sensory profiles of Italian monovarietal extra-virgin olive oils. J Sci Food Agric 90(15):2641–2648. https://doi.org/10.1002/jsfa.4133
Rudolf JD, Dong LB, Cao H, Hatzos-Skintges C, Osipiuk J, Endres M, Chang CY, Ma M, Babnigg G, Joachimiak A, Phillips GN Jr (2016) Structure of the ent-copalyl diphosphate synthase PtmT2 from Streptomyces platensis CB00739, a bacterial type II diterpene synthase. J Am Chem Soc 138(34):10905–10915. https://doi.org/10.1021/jacs.6b04317
Saleem M (2009) Lupeol, a novel anti-inflammatory and anti-cancer dietary triterpene. Cancer Let 285(2):109–115. https://doi.org/10.1016/j.canlet.2009.04.033
Salehi B, Upadhyay S, Erdogan OI, Kumar JA, Jayaweera LDS, Dias AD, Sharopov F, Taheri Y, Martins N, Baghalpour N, Cho CW (2019) Therapeutic potential of α-and β-pinene: a miracle gift of nature. Biomolecules 9(11):738. https://doi.org/10.3390/biom9110738
Sanz C, Perez AG, Olias R, Olias JM (1999) Quality of strawberries packed with perforated polypropylene. J Food Sci 64(4):748–752. https://doi.org/10.1111/j.1365-2621.1999.tb15124.x
Sarria S, Wong B, Martín HG, Keasling JD, Peralta-Yahya P (2014) Microbial synthesis of pinene. ACS Synth Biol 3(7):466–475. https://doi.org/10.1021/sb4001382
Schmelz EA, Huffaker A, Sims JW, Christensen SA, Lu X, Okada K, Peters RJ (2014) Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J 79(4):659–678. https://doi.org/10.1111/tpj.12436
Skrovankova S, Sumczynski D, Mlcek J, Jurikova T, Sochor J (2015) Bioactive compounds and antioxidant activity in different types of berries. Int J Mol Sci 16(10):24673–24706. https://doi.org/10.3390/ijms161024673
Takemura M, Tanaka R, Misawa N (2017) Pathway engineering for the production of β-amyrin and cycloartenol in Escherichia coli—a method to biosynthesize plant-derived triterpene skeletons in E. coli. Appl Microbiol Biotechnol 101(17):6615–6625. https://doi.org/10.1007/s00253-017-8409-z
Töpfer N, Fuchs LM, Aharoni A (2017) The PhytoClust tool for metabolic gene clusters discovery in plant genomes. Nucleic Acids Res 45(12):7049–7063. https://doi.org/10.1093/nar/gkx404
This P, Lacombe T, Thomas MR (2006) Historical origins and genetic diversity of wine grapes. Trends Genet 22(9):511–9. https://doi.org/10.1016/j.tig.2006.07.008
Van Der Donk WA (2015) Bacteria do it differently: an alternative path to squalene. ACS Cent Sci. https://doi.org/10.1021/acscentsci.5b00142
Vaughan MM, Christensen S, Schmelz EA, Huffaker A, Mcauslane HJ, Alborn HT, Romero M, Allen LH, Teal PE (2015) Accumulation of terpenoid phytoalexins in maize roots is associated with drought tolerance. Plant Cell Environ 38(11):2195–2207. https://doi.org/10.1111/pce.12482
Vespermann KA, Paulino BN, Barcelos M, Pessôa MG, Pastore GM, Molina G (2017) Biotransformation of α-and β-pinene into flavor compounds. Appl Microbiol Biotechnol 101(5):1805–1817. https://doi.org/10.1007/s00253-016-8066-7
Viet TD, Xuan TD, Anh LH (2021) α-Amyrin and β-amyrin Isolated from Celastrus hindsii leaves and their antioxidant, anti-xanthine oxidase, and anti-tyrosinase potentials. Molecules 26(23):7248. https://doi.org/10.3390/molecules26237248
Wang C, Gong X, Bo A, Zhang L, Zhang M, Zang E, Zhang C, Li M (2020) Iridoids: research advances in their phytochemistry, biological activities, and pharmacokinetics. Molecules 25(2):287. https://doi.org/10.3390/molecules25020287
Weston-Green K, Clunas H, Jimenez Naranjo C (2021) A review of the potential use of pinene and linalool as terpene-based medicines for brain health: discovering novel therapeutics in the flavours and fragrances of cannabis. Front Psychiat 12:1309. https://doi.org/10.3389/fpsyt.2021.583211
Wilderman PR, Peters RJ (2007) A single residue switch converts abietadiene synthase into a pimaradiene specific cyclase. J Am Chem Soc 129(51):15736–15737. https://doi.org/10.1021/ja074977g
Winnacker M (2018) Pinenes: abundant and renewable building blocks for a variety of sustainable polymers. Angew Chem Int Ed 57(44):14362–14371. https://doi.org/10.1002/anie.201804009
Xue Z, Duan L, Liu D, Guo J, Ge S, Dicks J, ÓMáille P, Osbourn A, Qi X (2012) Divergent evolution of oxidosqualene cyclases in plants. New Phytologist 193(4):1022–1038. https://doi.org/10.1111/j.1469-8137.2011.03997.x
Zerbe P, Rodriguez SM, Mafu S, Chiang A, Sandhu HK, O’Neil-Johnson M, Starks CM, Bohlmann J (2015) Exploring diterpene metabolism in non-model species: transcriptome-enabled discovery and functional characterization of labda-7, 13E-dienyl diphosphate synthase from Grindelia robusta. Plant J 83(5):783–793. https://doi.org/10.1111/tpj.12925
Zhang G, Cao Q, Liu J, Liu B, Li J, Li C (2015) Refactoring β-amyrin synthesis in Saccharomyces cerevisiae. AIChE J 61(10):3172–3179. https://doi.org/10.1002/aic.14950
Zhang Y, Wang Y, Xing J, Wan J, Wang X, Zhang J, Wang X, Li Z, Zhang M (2020) Copalyl diphosphate synthase mutation improved salt tolerance in maize (Zea mays. l) via enhancing vacuolar Na+ sequestration and maintaining ROS homeostasis. Front Plant Sci 11:457. https://doi.org/10.3389/fpls.2020.00457
Funding
No funding was received to assist with the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
SAA carried out the computational framework and analyses. SAA compiled the first draft of the manuscript and both authors contributed to writing the final version. KLF edited the manuscript before submission. Both authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Ethical approval
Not applicable
Consent for publication
Not applicable
Availability of data and materials
All necessary data supporting the conclusions of this article will be available by the authors without undue reservation.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Al-Salihi, S.A.A., Ford, K.L. Comparative bioinformatics analysis of the biosynthetic pathways and key candidate genes of three species, Vitis vinifera, Fragaria vesca and Olea europaea, furnish enzyme sets for the production of pharmaceutically valuable terpenes in heterologous hosts. J. Plant Biochem. Biotechnol. 32, 421–437 (2023). https://doi.org/10.1007/s13562-022-00823-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-022-00823-z