Abstract
Grapevine (Vitis vinifera) is widely applicated in food industry, which shows high economical and nutritional values. However, growth of grapevine was usually affected by various environmental stresses, such as salt, drought and disease. Ubiquitin fusion degradation protein 1 (UFD1) is an essential ubiquitin-recognition protein facilitates regulation of stress response through ERAD pathway. Even though, a comprehensive investigation of UFD1 genes in the plant species is still lacking. Here we identified three VvUFD1 proteins from genome of grapevine, which were assigned into different subgroups. All VvUFD1 genes contain highly conserved motifs in structure. Several cis-elements that related to fruit development and stress response were found in the promoter regions of VvUFD1 genes, including bHLH, NCA, MYB, HD-ZIP, GATA and AP2. Expression analysis found VvUFD1 genes showed different expression patterns in different tissues. Most importantly, VvUFD1 genes were found to be involved in salt stress response during growth of grapevine. Transcriptomic analyses were investigated for further understanding the genes’ function. Expression of VvUFD1 were increased at late stage of berry ripening. In addition, expression of VvUFD1 were also regulated by elevated light treatment and pathogen Neofusicoccum parvum infection. Co-expression network analysis revealed several major transcription factors that co-expressed with VvUFD1 genes. These results provide a basis for investigating the function of UFD1 genes in plant species and expand understanding of the regulation of berry development and salt stress response in grapevine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Grapevine (Vitis vinifera) is economically one of the most important perennial fruit crops in the world, which shows high nutritional value and wide application, such as wine production, consumed fresh, processed for juice and raisins (Myles et al. 2011). However, growth of grapevine was threatened by various biotic and abiotic stresses like drought, salinity and various diseases, thereby affecting fruit yield and quality (Keller 2010). Rapid ongoing global climate change is increasing the urgency to study the plant responses to environmental stresses. To improve the stress tolerance during grape development, there’s a high demand to identify key genes involved in stress and elucidate the regulation pathways under stress conditions.
The endoplasmic reticulum (ER) is one of the most functionally organelles in eukaryotic cells, which plays a central role in regulation of stress responses in both plant and animal cells (Schroder and Kaufman 2005). Extreme environments stresses usually lead to the increasing of unfolded proteins in cell, which afterwards cause endoplasmic reticulum (ER) stress (Lee 2001). Hence, it is critical for cell survival to remove these unfolded proteins through various quality control pathways through the ubiquitin proteasome system (Smalle and Vierstra 2004). Endoplasmic reticulum associated degradation (ERAD) is one of the protein quality control process that functions to removing abnormal proteins from ER. During ERAD, the target proteins could be firstly recognized by molecular chaperones and associated factors, translocated into the cytoplasm, and degraded by the ubiquitin–proteasome machinery (Vembar and Brodsky 2008). It has been well studied that a highly conserved chaperone complex Ufd1-Npl4-p97 plays a central role in the action of ERAD process, which contains Ufd1 (ubiquitin fusion degradation 1), Npl4 (nuclear protein localization homolog 4), and bound to a conserved ATPase (p97/VCP in mammals and Cdc48 in yeast) (Byrne et al. 2017; Bodnar et al. 2018). Misfolded proteins could be identified and ubiquitinated by the action of ubiquitin activating (E1), conjugating (E2), and ligase (E3) enzymes (Pickart 2001). Subsequently, the ubiquitin-tagged proteins could be bound by Ufd1-Npl4-p97 complex, extracted out and delivered to the proteasome for processing (Ye et al. 2001; Romisch 2005).
As the essential ubiquitin-recognition protein of ERAD pathway, the first UFD1 gene mutant was identified from yeast (Saccharomyces cerevisiae) with disturbed degradation process (Johnson et al. 1995). The conserved N domain of UFD1 has two distinct binding sites, which could be used for monoubiquitin and polyubiquitin, while a higher affinity for polyubiquitin was observed than monoubiquitin (Walters, 2005; Park et al., 2005). A population genetic study showed association between SNPs mutation of UFD1 and schizophrenia (Xie et al. 2008). Recent study suggested that expression of UFD1 could be regulated by ER stress to trigger cell cycle control, which contributes to ERAD in mammalian cells (Chen et al. 2011).
However, UFD1 members was barely reported in plant species compared to the abundance findings in yeast and mammals. At present, none of UFD1 genes was reported in grapevine. In order to identify UFD1 gene family and their functions in grapevine, the objectives of this study including: (1) screen UFD1 genes in grapevine genome using UFD1 domain; (2) determine gene structure and amino acid conserved functional domains of the grapevine UFD1, and analyzed the the cis-elements in their promoters; (3) investigate the transcriptional levels of UFD1 during fruit ripening, as also as response to various abiotic and biotic stresses. The results of this study could help us getting insight to the structure and functions of UFD1 genes in grapevine, which might have potential to be used in improvement of grapevine and other fruit trees.
Materials and methods
Plant materials and stress treatments
Grapevine rootstock cultivar ‘Kangzhen 5’ used in this study was derived from cross of ‘Beta’ (V. riparia × V. labrusca) and ‘420A’ (V. berlandieri × V. riparia), has high resistant to salinity, phylloxera and root-knot nematode. Cuttings of ‘Kangzhen 5’ were firstly rooted in humid sand crates, which were placed in a controlled culture room (25 °C, 90% humidity and 16/8 h day/night photoperiod) during dormancy. Young grapevine plantlets at 5 leaves separated stage were moved into pots containing soil-peat-sand (3:1:1) in a controlled greenhouse (24 °C and 16/8 h day/night photoperiod). After 2 months growth, salt treatment was applied using 150 mM NaCl solution. Fourth-unfolded leaves were collected at 0, 6, 24, 48 and 72 h after treatment. Roots, stems, leaves, petioles, tendrils, flowers at 14 leaves separated stage (EL18) and fruits at ripening stage (EL38) were sampled for tissue-specific expression analysis. Three independent biological replications were performed. All samples were immediately frozen in liquid nitrogen and then stored at − 80 °C until further analysis.
Identification of UFD1 genes identification and phylogenetic analysis
The UFD1 domain model (PF03152.14) was downloaded from Pfam database (http://pfam.xfam.org/), which was used to create a hidden Markov model (HMM) file using HMMER v3.0 package at default settings (http://hmmer.janelia.org/). All putative UFD1 genes were identified by screening against protein databases of grape (Vitis vinifera), Arabidopsis thaliana and rice (Oryza sativa) using HMMER v3.0. The identified sequences were further validated to in Pfam database. The candidate UFD1 genes were confirmed if both zinc finger-like and UFD1 domains were present.
MAFFT v7.313 software was used for multiple sequence analysis of UFD1 proteins. Phylogenetic tree was constructed using MEGA 7.0 software with a bootstrapped Neighbor-Joining (NJ) method. Molecular weight (MW), protein isoelectric point (PI) and GRAVY score were analyzed using ExPASy—ProtParam tool (https://web.expasy.org/protparam/). Subcellular locations were predicted on WoLF PSORT (https://wolfpsort.hgc.jp/).
Conserved motif distribution and gene structure analysis
MEME suit (version 5.1.1, http://meme-suite.org/) was used to analyze the conserved motif in amino acid sequences of UFD1 members in grape, Arabidopsis and rice. The default parameters were set for motif modification with maximum motif number of 15. Schematic diagrams of gene and motif structure were generated using TBtools (https://github.com/CJ-Chen/TBtools).
Analysis of the cis-elements in the promoters of UFD1 genes
The upstream of 2000 bp regions of grape UFD1 genes from the translation start site (TSS) were extracted from grape genome databases. The cis-elements of transcriptional factors binding sites were predicted using PlantRegMap tools (http://plantregmap.cbi.pku.edu.cn/binding_site_prediction.php). The positions of development-related and stress-responsive cis-elements in major fruits were showed on sketch maps draw by in-house python script.
Expression analysis in different organs and salt stress
Total RNA was extracted from grapevine samples and reverse transcribed into cDNA. For real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), the gene-specific primers for UFD1 genes listed in Supplementary file 1 were used to amplify short fragments of genes. GAPDH gene was used as housekeeping genes to normalize the expression of UFD1 genes. Amplification conditions were set as 95 °C for 3 min, 40 cycles of 95 °C for 5 s, 60 °C for 20 s, and 72 °C for 20 s. Following the PCR, a melting curve analysis was performed. Relative gene expression was calculated using 2−ΔΔCT method (Livak and Schmittgen 2001).
Expression analysis using RNA-Seq data
To explore the expression profiles of grapevine UFD1 genes under various conditions, RNA-seq data were downloaded from NCBI Gene Expression Omnibus (GEO) for grape ripening (GSE98923), treatments of elevated light (GSE98873), and pathogen Neofusicoccum parvum (GSE58653). The gene expression level was calculated by the FPKM (fragments per kilobase of exon per million fragments mapped) method.
Co-expression network analysis
To constructed co-expression network of grapevine UFD1 genes, RNA-seq data were downloaded from GEO, including 219 samples for grape ripening (GSE98923), and 23 samples for treatments of elevated light (GSE98873). Pearson correlation coefficient between pairwise genes were calculated to determine the similarity of gene expression. The transcription factors were selected as candidate co-expression genes at cutoff threshold of coefficient > 0.9 and Q ≤ 0.01. Co-expression network was constructed and visualized by Cytoscape software v3.5.1 (Shannon et al. 2003).
Results
Genome-wide identification of grape UFD1 genes
A total of three UFD1 genes were identified from grape genome by searching UFD1 domain in HMMER 3.0 platform. The three members were named as VvUFD1a, VvUFD1b and VvUFD1c, respectively and the corresponding identifiers are VIT_03s0038g00750, VIT_06s0004g05010 and VIT_12s0059g01100 in CRIBIv1 grape gene annotation database. VvUFD1a encodes a 309 amino acid (aa) protein, while VvUFD1b encodes a 165 aa protein and VvUFD1c encodes the longest 569 aa protein. The basic information of the three genes was listed in Table 1. Sequence of VvUFD1s were listed in Supplementary file 2.
Phylogenetic analysis, gene structure and motifs distribution of UFD1 proteins
To investigate the phylogenetic relationship between VvUFD1 proteins with their orthologous in other plant species, three Arabidopsis UFD1 proteins (AtUFD1a, AtUFD1b, AtUFD1c) and five rice UFD1 proteins (OsUFD1a, OsUFD1b, OsUFD1c, OsUFD1d, OsUFD1e) were isolated from their genomes. Sequences of Arabidopsis and rice UFD1 proteins were showed in Supplementary file 2. The result showed that VvUFD1a was classified close to OsUFD1a, AtUFD1a and AtUFD1c. VvUFD1c was in a distant subgroup together with OsUFD1c, while VvUFD1b was solely in a unique branch (Fig. 1). The phylogenetic information provides us a novel insight of evolutionary relationship of UFD1 genes among different plant species.
Phylogenetic analysis, gene structures and motifs distribution of UFD1 proteins from grapevine (VvUFD1), Arabidopsis thaliana (AtUFD1) and rice (OsUFD1). Phylogenetic tree was generated using the Neighbor-Joining method with 1000 bootstrap. The conserved motifs were analyzed using MEME program. Sequence of UFD1 proteins and motifs were showed in Supplementary file 2 and 3
We further investigated the gene structures of UFD1, which found genes classified in the same group usually showed a similar intro/exon composition. Among 11 UFD1 members, the number of coding sequence (CDS) regions varied from 3 to 9 (Fig. 1). There were 8 CDS in VvUFD1a, which distributed in similar pattern with OsUFD1a, AtUFD1a and AtUFD1c. Both VvUFD1c and OsUFD1c harbored 3 CDS. In addition, 4 CDS were found in VvUFD1b (Fig. 1).
Conserved motif composition of UFD1 proteins were investigated using MEME software. Among 15 conserved motifs, motif 1 was present in all UFD1 members across species, motif 2 and 3 were present in majority of UFD1 proteins except VvUFD1b (Fig. 1). VvUFD1b only included 3 conserved motifs due to its short sequence. For subgroup of VvUFD1c and OsUFD1c, there were motif 9, 10, 11, 12 and 14 that uniquely within subgroup (Fig. 1). These results could enhance the understanding of similarity and diversity of function of UFD1 proteins.
Analysis of cis-elements in Promoter of VvUFD1 Genes
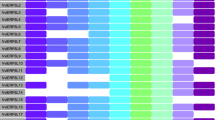
Cis-elements are part of promoter regions that regulate the transcription of neighboring genes, which are vital components of genetic regulatory networks. In order to get insight into the transcription regulation of VvUFD1 genes, we investigated the distribution of cis-elements in the promoters of these genes. As showed in Fig. 2, 20 different type of elements were found in promoters of three VvUFD1 genes, including 7 types in VvUFD1a, 14 types in both VvUFD1b and VvUFD1c. Among them, one element (LBD) only present in VvUFD1a promoter, while three elements (BES1, bHLH, TALE) only found in VvUFD1b promoter, and four elements (MYB_related, NAC, TCP, Trihelix) were uniquely harbored in promoter of VvUFD1c. In addition, a portion of cis-elements were known to regulate fruit development and stress response, such as bHLH, NCA, MYB, HD-ZIP, GATA. AP2 transcription factor, whose binding site was found in all three VvUFD1 promoters, also has been reported to be involved in fruit ripening through regulation of ethylene signaling pathway.
Expression analysis of VvUFD1 genes in different organs and salt stress
The expression profiles of VvUFD1 genes in different organs and salt stress were investigated using qPCR analysis. The results suggested that expression of VvUFD1a showed higher levels in petiole and stem, and lowest level in root (Fig. 3a). However, VvUFD1c exhibited higher transcript abundance in root and fruit, and relatively lower levels in stem, leave, petiole, flower and tendril (Fig. 3c). The results also determined that VvUFD1 genes showed different response patterns to salt stress in leaves samples. Expression of VvUFD1a was slightly decreased at early stage (6 h), then gradually increased along the duration of stress, and showed peak level (6.65 ×) at 48 h after stress (Fig. 3b). Conversely, expression of VvUFD1c was slightly up-regulated at 6 h, and sharply reduced at 24, 48 and 72 h by salt stress compared with pre-treatment (Fig. 3d).
Expression profiles of VvUFD1 genes in different organs of grapevine and under salt stress. a Expression levels of VvUFD1a in root, stem, leave, petiole, flower, tendril and fruit. b Expression pattern of VvUFD1a in leaves after salt treatment at 6, 24, 48 and 72 h. c Expression levels of VvUFD1c in root, stem, leave, petiole, flower, tendril and fruit. d Expression pattern of VvUFD1c in leaves after salt treatment at 6, 24, 48 and 72 h
Expression analysis of the VvUFD1 genes using RNA-Seq data
To analyze the expression of VvUFD1 genes during grapevine berry development and ripening, RNA-Seq sequencing data of grapevine berries were downloaded from the SRA database (GSE98923) and used to calculate gene expression levels. Berry samples were collected at 10-d intervals or weekly (0–12) from fruit-set to maturity in grapevine genotypes “Cabernet Sauvignon” for three consecutive years (2012, 2013, 2014) (Fasoli et al. 2018). As showed in Fig. 4, expression of VvUFD1a and VvUFD1c were primarily increased throughout development process and reached highest expression level at the late-ripening stages. Comparatively, expression of VvUFD1b was slightly increased at the earlier sample points but down-regulated around mid-development, while rapidly increased at the late-ripening stage. It was also noticed that VvUFD1a and VvUFD1c showed much higher transcript accumulation (higher FPKM value) than VvUFD1b during berry development (Fig. 4).
Expression pattern of VvUFD1 genes during grapevine berry development and ripening. Grapevine (Vitis vinifera) Cabernet Sauvignon was used in this study. Berries were collected at 10-d intervals in 2012 and weekly in 2013 and 2014, beginning at fruit set and continuing until harvest (Fasoli et al. 2018)
We also investigated the expression profiles of grapevine VvUFD1 genes in response to light exposure treatment at berry development stage using transcriptome sequencing data (GSE98873). Briefly, elevated light exposure was applied by leaf-removal treatment, and the berries were sampled at green (pea-sized) (EL31), pre-veraison (EL33), veraison (EL35), and the ripe-stage (EL38) (du Plessis et al. 2017). VvUFD1a and VvUFD1c showed much higher expression levels (higher FPKM value) than VvUFD1b induced by light treatment (Fig. 5). Compared to control panels, elevated light treatment increased expression of VvUFD1c at EL31, EL33 and EL34, but did not show obvious difference at late stage (EL38). Reversely, expression of VvUFD1b was down-regulated at EL33 by elevated light, but highly up-regulated at EL34 and EL38. However, expression of VvUFD1a was not obviously affected by elevated light treatment (Fig. 5).
Expression pattern of VvUFD1 genes in response to elevated light exposure at four grape phenological stages (EL31, EL33, EL34, EL 38) (du Plessis et al. 2017)
Neofusicoccum parvum infects the wood of grapevines and other horticultural crops, killing the fruit-bearing shoots (Czemmel et al. 2015). We examined expression profile of VvUFD1 genes at inoculated (IW) and non-inoculated (NIW) plants using RNA-Seq dataset (GSE58653) (Czemmel et al. 2015). Expression of VvUFD1a did not change in non-inoculated plants, but highly increased following inoculation of Neofusicoccum parvum (Fig. 6). Under non-inoculation condition, expression of VvUFD1b was decreased after 0.5–1.5 months. However, the expression was inversely up-regulated in inoculation group (Fig. 6). In addition, expression of VvUFD1c were slightly increased in both NIW and IW groups (Fig. 6).
Expression pattern of VvUFD1 genes in response to infection of pathogen Neofusicoccum parvum. Leaf samples were collected from both inoculated-wounded (IW) and non-inoculated-wounded (NIW) plants at 0, 0.5, 1, 1.5 months post-inoculation (MPI). Samples from 0.5, 1 and 1.5 MPI were pooled to compare versus sample at 0 MPI (Czemmel et al. 2015)
Co-expression network of the VvUFD1 Genes
The transcriptome datasets conferring grape berry ripening and elevated light stress were used to search co-expression relationship between the VvUFD1 genes and major transcription factors. There were 9 pairs positive co-expression and 16 pairs negative co-expression were detected for VvUFD1a, including 4 MYB genes, 3 bHLH genes, 3 Dof genes, 3 Trihelix genes, 3 TCP genes and several other genes (Fig. 7). For VvUFD1c, there were 35 pairs positive co-expression and 7 pairs negative co-expression were detected, and the major co-expression genes containing 5 NAC genes, 5 MYB genes, 3 bHLH genes, and 3 bZIP genes (Fig. 7). In addition, several TF genes were overlapped in the two networks of VvUFD1a and VvUFD1c, including bHLH, LBD, MYB, WRKY, NAC, Trihelix, bZIP, Dof and TCP genes (Fig. 7).
Discussion
Identification of UFD1 genes in plant species
In this study, three VvUFD1 family genes were identified from genome of grapevine (Table 1). Moreover, structure and motif analysis demonstrated that VvUFD1 proteins had several highly conserved domains with the orthologous in Arabidopsis and rice genomes (Fig. 1), which imply a strong evolutionary conservation of UFD1 proteins. UFD1 protein was first identified in yeast (Johnson et al. 1995), and then isolated from other eukaryotes, such as human (Pizzuti et al. 1997), mouse (Botta et al. 1997), Gallus gallus, Xenopus laevis and Drosophila melanogaster (Ratti et al. 2001). However, only few of UFD1-like proteins were isolated and function analyzed from plants species until recently, although they have been predicted from genome sequence of many plants. Wei et al. (2009) reported two UFD1 paralog proteins from wheat, which showed highly evolutionarily conserved ubiquitin-binding domain. In addition, another UFD1 gene was cloned from tomato (Lai et al. 2012). In addition to UFD1 genes in grapevine, we also identified three Arabidopsis UFD1 proteins (AtUFD1a, AtUFD1b, AtUFD1c) and five rice UFD1 proteins (OsUFD1a, OsUFD1b, OsUFD1c, OsUFD1d, OsUFD1e) by searching UFD1 domains in their genomes (Fig. 1). All these results indicate that UFD1 is a low copy gene family. There is only one UFD1 gene copy in genome of yeast and mammals separately (Johnson et al. 1995; Pizzuti et al. 1997; Botta et al. 1997). The higher gene copy in plant species might be caused by genome or segment duplication during species evolution, which could make them better adapted to adverse growth environments.
The VvUFD1 genes might play roles in fruit development and ripening
Expression of VvUFD1 genes were found highly increased at the late-ripening stage of grape berry (Fig. 4), which suggest VvUFD1 genes might play important roles during berry development and ripening. It has been reported that organic acids, tannins, hydroxycinnamates, phenolic precursors and sugars were accumulated during the process of berry development, while organic acids and synthesis of volatile aromas were lost (Conde et al. 2007). It was known that hormones play central roles in regulation of above metabolism process during berry development. Ethylene and abscisic acid (ABA) induce ripening, whereas auxin indole-3-acetic acid inhibits ripening (Davies and Bottcher 2009). Our study showed evidence that VvUFD1 genes might be involved in regulation of ethylene pathway. We found that several AP2 transcription factor binding site in all three VvUFD1 promoters (Fig. 2). AP2 is an ethylene-responsive factor that has been reported to be involved in grape ripening through regulation of ethylene signaling pathway (Licausi et al. 2010). A bHLH binding site was found in promoter of VvUFD1b (Fig. 2). SlbHLH95 also was proved to be participated in the regulation of fruit ripening through ethylene metabolism pathway (Zhang et al. 2020). In addition, co-expression network analysis found that several transcription factors including NAC, WRKY and bHLH showed co-expression relationship with VvUFD1 genes, suggesting they might be regulators of the expression of the VvUFD1 genes (Fig. 7). Recently, VvibHLH075 and VviWRKY19 were proved to play key roles in regulation transition from immature to mature grape berry development (Palumbo et al. 2014). In another study, the expression of VvibHLH075 and VviWRKY19 were found to be up-regulated during the grape ripening, which could be used as positive biomarkers of fruit ripening (Fasoli et al. 2018). In addition, bHLH075 and WRKY19 could also regulated the expression of several NAC transcription factor genes, which were involved in regulation of plant development in many plant species (Raman et al. 2008; Fabi et al. 2012; Wang et al. 2013). It is interesting that the NAC binding cis-elements were also found in the promoter region of VvUFD1 genes, and NAC genes were identified as co-repression genes of VvUFD1 genes in our study (Fig. 2 and 7). All these findings implied that VvUFD1 genes might be involved in grape ripening through activation of a series genes.
The expression of VvUFD1 genes were regulated by environmental stresses
It has been determined that UFD1 could involves in degradation of abnormal proteins and thus helps to restore endoplasmic reticulum protein homeostasis through endoplasmic reticulum (ER)-associated degradation (ERAD) process (Ye et al. 2001; Kim et al. 2015). Compared to abundance evidence in human and mouse studies, few UFD1 has been reported to be involved in stress response in plant species. In our study, we found that VvUFD1 genes could be regulated by various abiotic and biotic stresses, including salt stress (Fig. 3), elevated light treatment (Fig. 5) and pathogen Neofusicoccum parvum (Fig. 6). Although three VvUFD1 members exhibit different response patterns to those stresses. Lai et al. (2012) demonstrated that silencing UFD1 in Nicotiana benthamiana could significantly alleviating the necrotic symptoms of tobacco wild fire disease. Recently, a possible degradation pathway was reported in wheat that TaPI4KIIγ could interacts with TaUFD1 bound to ubiquitin, and then the protein complex is transported to the 26S proteasome degradation system, which has been found plays important roles in regulation of abiotic and biotic stress responses in plants (Dielen et al. 2010; Liu et al. 2013). However, it is still not clear the pathways of VvUFD1 genes involved in stress response, further studies are needed to explore the regulation mechanisms.
Conclusion
In conclusion, three VvUFD1 genes were identified from genome of grapevine. All VvUFD1 members contain highly conserved motifs. Several cis-elements related to fruit development and stress response were found in the promoter regions of VvUFD1 genes. Expression of VvUFD1 genes were regulated by salt stress, elevated light treatment and pathogen Neofusicoccum parvum infection. In addition, expression of VvUFD1 were increased at late stage of berry ripening. These results suggesting VvUFD1 genes may be involved in the regulation of fruit development and stress response. The findings in this study will enhance our understanding of the structure and function of UFD1 family genes, and allow for exploring further application in breeding of grapevine.
Abbreviations
- UFD1:
-
Ubiquitin fusion degradation protein 1
- ER:
-
Endoplasmic reticulum
- ERAD:
-
Endoplasmic reticulum associated degradation
- MW:
-
Molecular weight
- PI:
-
Protein isoelectric point
- TSS:
-
Translation start site
- qRT-PCR:
-
Quantitative reverse transcriptase polymerase chain reaction
- CDS:
-
Coding sequence
References
Araki E, Oyadomari S, Mori M (2003) Endoplasmic reticulum stress and diabetes mellitus. Intern Med 42:7–14
Bodnar NO, Kim KH, Ji Z, Wales TE, Svetlov V, Nudler E, Engen JR, Walz T, Rapoport TA (2018) Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4. Nat Struct Mol Biol 25(7):616–622
Botta A, Jurecic V, Pizzuti A, Novelli G, Dallapiccola B, Baldini A (1997) Assignment of the gene for a ubiquitin fusion degradation protein (UFD1) to mouse chromosome 16B1-B4, syntenic with the Tuple1 gene. Cytogenet Cell Genet 77:264–265
Byrne DJ, Harmon MJ, Simpson JC, BlackstoneOSullivan CNC (2017) Roles for the VCP co-factors Npl4 and Ufd1 in neuronal function in Drosophila melanogaster. J Genet Genom 44(10):493–501
Chen M, Gutierrez GJ, Ronai ZA (2011) Ubiquitin-recognition protein Ufd1 couples the endoplasmic reticulum (ER) stress response to cell cycle control. Proc Natl Acad Sci 108(22):9119–9124
Conde C, Silva P, Fontes N, Dias ACP, Tavares RM, Sousa MJ, Agasse A, Delrot S, Geros H (2007) Biochemical changes throughout grape berry development and fruit and wine quality. Food 1:1–22
Czemmel S, Galarneau ER, Travadon R, McElrone AJ, Cramer GR, Baumgartner K (2015) Genes expressed in grapevine leaves reveal latent wood infection by the fungal pathogen Neofusicoccum parvum. PLoS ONE 10(3):e0121828
Davies C, Bottcher C (2009) Hormonal control of grape berry ripening. In: Roubelakis-Angelakis KA (ed) Grapevine molecular physiology & biotechnology, 2. Springer, Dordrecht, pp 229–261
Dielen AS, Badaoui S, Candresse T, German-Retana S (2010) The ubiquitin/26S proteasome system in plant–pathogen interactions: a never-ending hide-and-seek game. Mol Plant Pathol 11:293–308
du Plessis K, Young PR, Eyéghé-Bickong HA, Vivier MA (2017) The transcriptional responses and metabolic consequences of acclimation to elevated light exposure in grapevine berries. Front Plant Sci 8:1261
El-Gebali S, Mistry J, Bateman A et al (2019) The Pfam protein families database in 2019. Nucleic Acids Res 47(D1):D427–D432
Fabi JP, Seymour GB, Graham NS, Broadley MR, May ST, Lajolo FM, Cordenunsi BR, Oliveira do Nascimento JR (2012) Analysis of ripening-related gene expression in papaya using an Arabidopsis-based microarray. BMC Plant Biol 12:242
Fasoli M, Richter CL, Zenoni S, Bertini E, Vitulo N, Dal Santo S, Dokoozlian N, Pezzotti M, Tornielli GB (2018) Timing and order of the molecular events marking the onset of berry ripening in grapevine. Plant Physiol 178(3):1187–1206
Huiting LN, Samaha Y, Zhang GL, Roderick JE, Li B, Anderson NM, Wang YW, Wang L, Laroche F, Choi JW, Liu CT, Kelliher MA, Feng H (2018) UFD1 contributes to MYC-mediated leukemia aggressiveness through suppression of the proapoptotic unfolded protein response. Leukemia 32(11):2339–2351
Johnson ES, Bartel B, Seufert W, Varshavsky A (1992) Ubiquitin as a degradation signal. EMBO J 11:497–505
Johnson ES, Ma PC, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270:17442–17456
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780
Keller M (2010) Managing grapevines to optimise fruit development in a challenging environment: a climate change primer for viticulturists. Aust J Grape Wine Res 16:56–69
Kim H, Bhattacharya A, Qi L (2015) Endoplasmic reticulum quality control in cancer: friend or foe. Semin Cancer Biol 33:25–33
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Lai YY, Li F, Xu YP, Cai XZ (2012) Functional analysis of a ubiquitin fusion degradation protein gene UFD1 in regulation of plant disease and stress resistance. J Zhejiang Univ 38:21–27
Lee AS (2001) The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci 26:504–510
Licausi F, Giorgi FM, Zenoni S, Osti F, Pezzotti M, Perata P (2010) Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genom 11:719
Liu P, Xu ZS, Pan-Pan L, Hu D, Chen M, Li L, Ma Y (2013) A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. J Exp Bot 64(10):2915–2927
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Myles S, Boyko AR, Owens CL, Brown PJ, Grassi F, Aradhya MK, Prins B, Reynolds A, Chia J-M, Ware D (2011) Genetic structure and domestication history of the grape. Proc Natl Acad Sci USA 108:3530–3535
Palumbo MC, Zenoni S, Fasoli M, Massonnet M, Farina L, Castiglione F, Pezzotti M, Paci P (2014) Integrated network analysis identifies fight-club nodes as a class of hubs encompassing key putative switch genes that induce major transcriptome reprogramming during grapevine development. Plant Cell 26:4617–4635
Park S, Isaacson R, Kim HT, Silver PA, Wagner G (2005) Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure 13:995–1005
Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533
Pizzuti A, Novelli G, Ratti A, Amati F, Mari A, Calabrese G, Nicolis S, Silani V, Marino B, Scarlato G, Ottolenghi S, Dallapiccola B (1997) UFD1L, a developmentally expressed ubiquitination gene, is deleted in CATCH 22 syndrome. Hum Mol Genet 6(2):259–265
Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K (2008) Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55:65–76
Ratti A, Amati M, Bozzali E (2001) Cloning and molecular characterization of three ubiquitin fusion degradation 1 (UFD1) ortholog genesfrom Xenopuslaevis, Gallus gallus and Drosophila melanogaster. Cytogenet Cell Genet 92:279–282
Romisch K (2005) Endoplasmic reticulum-associated degradation. Annu Rev Cell Dev Biol 21:435–456
Schroder M, Kaufman RJ (2005) The mammalian unfolded protein response. Annu Rev Biochem 74:739–789
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55:555–590
Vembar S, Brodsky J (2008) One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9:944–957
Walters KJ (2005) Ufd1 exhibits dual ubiquitin binding modes. Structure 13:943–944
Wang N, Zheng Y, Xin H, Fang L, Li S (2013) Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep 32:61–75
Wei L, Tao Y, Jia H, Zhang L, Xu P, Wang Y, Zhang ZH, Zhang C, Ma ZH (2009) Highly conserved UFD1 proteins among eukaryotes exhibit considerable C-terminus diversity in different taxa. Plant Mol Biol Rep 27:439–447
Xie L, Ye L, Ju G, Xu Q, Zhang X, Liu S, Shi J, Yu Y, Wang Z, Shen Y, Wei J (2008) A family- and population-based study of the UFD1L gene for schizophrenia. Am J Med Genet B Neuropsychiatr Genet 147B(7):1076–1079
Ye Y, Meyer HH, Rapoport TA (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from ER into the cytosol. Nature 414:652–656
Zhang L, Kang J, Xie Q, Gong J, Shen H, Chen Y, Chen G, Hu Z (2020) The basic helix-loop-helix transcription factor bHLH95 affects fruit ripening and multiple metabolisms in tomato. J Exp Bot 71(20):6311–6327
Acknowledgements
We gratefully thank the funding from the National Natural Science Foundation of China (Youth Fund Project, No. 31301751), China Agriculture Research System of MOF and MARA (CARS-29-13) and the Key Project for New Agricultural Cultivar Breeding in Zhejiang Province under Grant 2021C02066-6.
Author information
Authors and Affiliations
Contributions
LW and JW design the experiments, analyzed and interpreted the data and was a major contributor in writing the manuscript. JC and JX conducted experiments and analyzed the data. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13562_2021_742_MOESM1_ESM.xlsx
Supplementary file 4: Transcriptomic data of grape ripening, treatments of elevated light, and pathogen Neofusicoccum parvum. (XLSX 14 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wei, L., Cheng, J., Xiang, J. et al. Genome-wide identification and characterization of grapevine UFD1 genes during berry development and salt stress response. J. Plant Biochem. Biotechnol. 31, 592–601 (2022). https://doi.org/10.1007/s13562-021-00742-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-021-00742-5