Abstract
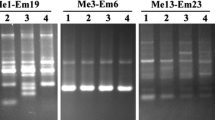
Amplified fragment length polymorphism (AFLP) analysis was conducted to investigate the level of polymorphism in four jute genotypes including two genotypes (JRC 321 and CMU 010) of Corchorus capsularis (the white jute) and two genotypes (JRO 524 and PPO4) of Corchorus olitorius (the tossa jute). Out of 1024 primer combinations that were tried, as many as 281 combinations of selective primers (13 EcoRI and 64 MseI) were selected, which produced a total of 9092 amplicons, including 752 (8.3%) polymorphic bands in C. capsularis and a total of 8856 amplicons including 1477 (16.7%) polymorphic bands in C. olitorius. The average number of bands/primer combination was 32.3 for C. capsularis and 31.5 for C. olitorius. For C. capsularis, highest polymorphism of 56.6% was shown by primer combination E35M50, while for C. olitorius highest polymorphism of 50% was shown by E41M91. In C. olitorius, 30–50% polymorphism was observed with 27 primer combinations, but in C. capsularis only 3 primer combinations gave this level of polymorphism. Similarly, in C. capsularis <10% polymorphism was detected by 115 primer combinations while in C. olitorius, <10% polymorphism was shown by only 56 primer combinations. These results indicate a higher level of polymorphism in C. olitorius relative to that in C. capsularis. The occurrence of such a large number of polymorphic AFLP markers will facilitate preparation of molecular maps and QTL analysis in jute.




Similar content being viewed by others
Abbreviations
- AFLP:
-
Amplified Fragment Length Polymorphism
- CMU:
-
Capsularis Mutant
- CTAB:
-
Cetyl Trimethyl Ammonium Bromide
- DTT:
-
Dithiothreitol
- JRC:
-
Jute Research Capsularis
- JRO:
-
Jute Research Olitorius
- PAGE:
-
Polyacrylamide Gel Electrophoresis
- PPO4:
-
Plant Physiology Olitorius 4
- QTL:
-
Quantitative Trait loci
- TBE:
-
Tris Borate EDTA
- TE:
-
Tris EDTA
References
Adawy SS, Assem SK, Hussein EHA, El-ltriby HA (2004) Development of AFLP markers and genotyping of elite maize inbred lines. Arab J Biotech 7:53–64
Adawy SS, Saker MM, Haggag WM, El-ltriby HA (2008) Amplified Fragment Length Polymorphism (AFLP) based molecular analysis of Egyptian barley lines and landraces differing in their resistance and susceptibility to leaf rust and net blotch diseases. Paper presented at the workshops “Better Plants for Better Life” conducted during the German/Egyptian Year of Science and Technology 2007 at ARC in Cairo, Egypt and FAL
Basu A, Ghosh M, Meyer R, Powell W, Basak SL, Sen SK (2004) Analysis of genetic diversity in cultivated jute determined by means of SSR markers and AFLP profiling. Crop Sci 44:678–685
Benor S, Blattner FR, Demissew S, Hammer K (2009) Collection and ethnobotanical investigation of Corchorus species in Ethiopia: potential leafy vegetables for dry regions. Genet Resour Crop Evol doi:10.1007/s10722-009-9470-y
Cho YG, Blair MW, Panaud O, McCouch SR (1996) Cloning and mapping of variety specific rice genomic DNA sequences: amplified fragment-length polymorphism (AFLP) from silver stained polyacrylamide gels. Genome 39:373–378
Ferriol M, Pico B, Nuez N (2003) Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers. Theor Appl Genet 107:271–282
Grot G, van Eeuwijk FA (2010) Codominant scoring of AFLP in association panels. Theor Appl Genet 121:337–351
Heywood VH, Brummitt RK, Culham A, Seberg O (2007) Flowering plant families of the world. Royal Botanical Gardens, Kew
Hill M, Witsenboe H, Zabeau M, Vos P, Kesseli R, Michelmore R (1996) PCR-based fingerprinting using AFLP’s as a tool for studying genetic relationships in Lactuca spp. Theor Appl Genet 93:1202–1210
Hossain MB, Awal A, Rahman MA, Haque S, Khan H (2003) Distinction between cold-sensitive and –tolerant jute by DNA polymorphisms. J Biochem Mol Biol 36:427–432
Keim P, Schupp JM, Travis SE, Clayton K, Zhu T, Shi T, Ferreira A, Webb DM (1997) A high-density soybean genetic map based on AFLP markers. Crop Sci 37:537–543
Lombard V, Baril CP, Dubreuil P, Blouet F, Zhang D (2000) Genetic relationship and fingerprinting of rapeseed cultivars by AFLP: Consequences for varietal registration. Crop Sci 40:1417–1425
Meksem K, Leister D, Peleman J, Zabeau M, Salamini F, Gebhardt C (1995) A high resolution map of the vicinity of the R1 locus on chromosome V of potato based on RFLP and AFLP markers. Mol Gen Genet 249:74–81
Mir RR, Rustgi S, Sharma S, Singh R, Goyal A, Kumar J, Gaur A, Tyagi AK, Khan H, Sinha MK, Balyan HS, Gupta PK (2008) A preliminary genetic analysis for fibre traits and the use of new genomic SSRs for genetic diversity in jute. Euphytica 161:413–427
Mir RR, Banerjee S, Das M, Gupta V, Tyagi AK, Sinha MK, Balyan HS, Gupta PK (2009) Development and characterization of large-scale simple sequence repeats in jute. Crop Sci 49:1687–1694
Qi X, Stam P, Lindhout P (1998) Use of locus specific AFLP markers to construct a high-density map in barley. Theor Appl Genet 96:376–384
Roy JK, Lakshmikumaran MS, Balyan HS, Gupta PK (2004) AFLP-based genetic diversity and its comparison with diversity based on SSR, SAMPL, and phenotypic traits in bread wheat. Biochem Genet 42:1–2
Roy A, Bandyopadhyay A, Mahaptra AK, Ghosh SK, Singh NK, Bansal KC, Koundal KR, Mohapatra T (2006) Evaluation of genetic diversity in jute (Corchorous species) using STMS, ISSR and RAPD markers. Plant Breed 125:292–297
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Santos CAF, Simon PW (2002) Some AFLP amplicons are highly conserved DNA sequences mapping to the same linkage group in two F2 populations of carrot. Genet Mol Biol 25:195–201
Tegelstrom H (1992) Detection of mitochondrial DNA fragments. In: Hoelzl AR (ed) Molecular genetic analysis of populations: a practical approach. IRL, Oxford, pp 89–114
Thomas CM, Vos P, Zabeau M (1995) Identification of amplified restriction fragment polymorphism (AFLP) markers tightly linked to the tomato Cf-9 gene for resistance to Cladosporium fulvum. Plant J 8:785–794
Varshney A, Mohapatra T, Sharma RP (2004) Development and validation of CAPS and AFLP markers for white rust resistance gene in Brassica junicea. Theor Appl Genet 109:153–159
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23(21):4407–4414
Waugh R, Bonar N, Baird E, Thomas B, Garner A, Hayes P, Powell W (1997) Homology of AFLP products in three mapping populations of barley. Mol Gen Genet 255:311–321
Acknowledgements
The financial support from the Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India is gratefully acknowledged. Department of Science and Technology (DST), Government of India, under its DST-FIST programme, provided some of the equipments used in this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Das, M., Banerjee, S., Topdar, N. et al. Development of large-scale AFLP markers in jute. J. Plant Biochem. Biotechnol. 20, 270–275 (2011). https://doi.org/10.1007/s13562-011-0058-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-011-0058-1