Abstract
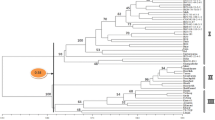
AFLP markers were evaluated for determining the phylogenetic relationships Lactuca spp. Genetic distances based on AFLP data were estimated for 44 morphologically diverse lines of cultivated L. sativa and 13 accessions of the wild species L. serriola, L. saligna, L. virosa, L. perennis, and L. indica. The same genotypes were analyzed as in a previous study that had utilized RFLP markers. The phenetic tree based on AFLP data was consistent with known taxonomic relationships and similar to a tree developed with RFLP data. The genetic distance matrices derived from AFLP and RFLP data were compared using least squares regression analysis and, for the cultivar data, by principal component analysis. There was also a positive linear relationship between distance estimates based on AFLP data and kinship coefficients calculated from pedigree data. AFLPs represent reliable PCR-based markers for studies of genetic relationships at a variety of taxonomic levels.
Similar content being viewed by others
References
Akopyanz N, Bukanov NO, Westblom TU, Berg DE (1992) PCR-based RFLP analysis of DNA sequence diversity in the gastric pathogen Helicobacter pylori. Nucleic Acids Res 20:6221–6225
Bonierbale MW, Plaisted RL, Tanksley SD (1988) RFLP maps based on a common set of clones reveal modes of chromosome evolution in potato and tomato. Genetics 120:1095–1103
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment lenght polymorphisms. Am J Hum Gen 32:314–331
Chalmers KJ, Waugh R, Sprent JI, Simons AJ, Powell W (1992) Detection of genetic variation between and within populations of Gliricidia sepium and G. maculata using RAPD markers. Genetics 69:465–472
Cox TS, Yiang YT, Gorman MB, Rogers DM (1985) Relationship between coefficient of parentage and genetic similarity indices in soybean. Crop Sci 25:529–532.
Cox TS, Murphy JP, Rodgers DM (1986) Changes in genetic diversity in the red winter wheat regions of the United States. Proc Natl Acad Sci USA, 83:5583–5586
Demeke T, Adams RP, Chibbar R (1992) Potential taxonomic use of random amplified polymorphic DNA (RAPD): a case study in Brassica. Theor Appl Genet 84:990–994
dos Santos JB, Nienhuis J, Skroch P, Tivang J, Slocum MK (1994) Comparison of RAPD and RFLP genetic markers in determining genetic similarity among Brassica oleracea L. genotype. Theor Appl Genet 87:909–915
Farrara BF, Ilott TW, Michelmore RW (1987) Genetic analysis of factors for resistance to downy mildew (Bremia lactucae) in species of lettuce (Lactuca sativa and L. serriola). Plant Pathol 36:499–514
Fukuoka S, Hosoka K, Kamijima O (1992) Use of random amplified polymorphic DNAs (RAPDs) for identification of rice accessions. Jpn J Genet 67:243–252
Gower JC (1966) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338
Heun M, Murphy JP, Phillips TD (1994) A comparison of RAPD and isozyme analyses for determining the genetic relationships among Avena sterilis L. accessions. Theor Appl Genet 87:689–696
Hulbert SH, Michelmore RW (1985) Linkage analysis of genes for resistance to downy mildew (Bremia lactucae) in lettuce (Lactuca sativa). Theor Appl Genet 70:520–528
Kazan K, Manners JM, Cameron DF (1993) Genetic relationships and variation in the Stylosanthes guianensis species complex assessed by random amplified polymorphic DNA. Genome 36:43–49
Kesseli RV, Michelmore RW (1986) Genetic variation and phylogenies detected from isozyme markers in species of Lactuca. J Hered 77:324–331
Kesseli R, Ochoa O, Michelmore R (1991) Variation at RFLP loci in Lactuca spp. and origin of cultivated lettuce (Lactuca sativa). Genome 34:430–436
Kesseli RV, Paran I, Michelmore RW (1994) Analysis of a detailed genetic linkage map of Lactuca sativa (lettuce) constructed from RAPD Markers. Genetics 136:1435–1446
Koller B, Lehmann A, Gessler C (1993) Identification of apple cultivars using RAPD markers. Theor Appl Genet 85:901–904
Kresovich S, Williams JGK, McFerson JR, Routman EJ, Schaal BA (1992) Characterization of genetic identities and relationships of Brassica oleracea L. via a random amplified polymorphic DNA assay. Theor Appl Genet 85:190–196
Lamboy WF (1994) Computing genetic similarity coefficients from RAPD data: the effects of PCR artifacts. PCR Meth Applic 4:31–37
Landry BS, Kesseli R, Leung H, Michelmore RW (1987) Comparison of restriction endonucleases and sources of probes for their efficiency in detecting restriction fragment length polymorphisms in lettuce (Lactuca sativa L.). Theor Appl Genet 74:646–653
Linquist K (1960) On the origin of cultivated lettuce. Hereditas 46:19–350
Martin JM, Blake TK, Hockett EA (1991) Diversity among North American spring barley cultivars based on coefficients of parentage. Crop Sci 31:1131–1137
Melchinger AE, Messmer MM, Lee M, Woodman WL, Lamkey KR (1991) Diversity and relationship among US maize inbreds revealed by restriction fragment length polymorphisms. Crop Sci 31:669–678
Michelmore RW, Anderson P, Okubara P, Witsenboer H (1994) Clusters of resistance genes in lettuce: map-based cloning in non-intensively studied species, In: Coruzzi G, Puigdomenech P (eds) NATO ASI series, vol H 81: plant molecular biology. Springer, Berlin, Hiedelberg New York, pp 501–509
Nei M, Li WH (1979) Mathematical models for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci, USA 76:5269–5273
Robinson RW, McCreight JD, Ryder EJ (1976) The genes of lettuce and closely related species. Plant Breed Rev 1:267–293
Rohlf FJ (1992) NTSYS-pc. Numerical taxonomy and multivariate analysis system. Version 1.70. Exeter Software, Setauket, N.Y.
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a laboratory manual, 2nd edn. Cold Spring Harbour Press, Cold Spring Harbour, N.Y.
Skroch P, Tivang J, Nienhuis J (1993) Analysis of genetic relationships using RAPD marker data. Bio/Technology 11:26–30
Smith OS, Smith JSC, Bowen SL, Tenborg RA, Wall SJ (1990) Similarities among a groups of elite maize inbreds as measured by pedigree F1 grain yield, heterosis, and RFLPs. Theor Appl Genet 80:833–840
Song K, Osborn TC, Williams PH (1990) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs) 3. Genome relationships in Brassica and related genera and the origin of B. oleracea and B. rapa (syn. campestris). Theor Appl Genet 79:497–506
Tanksley SD (1989) RFLP mapping in plant breeding: new tools for an old science. Biotechnology 7:257–264
Tanksley SD, Bernatzky R, Lapitan NL, Prince JP (1988) Conservation of gene repertoire but not gene order in pepper and tomato. Proc Natl Acad Sci USA 85:6419–6423
Tinker NA. Fortin MG, Mather DE (1993) Random polymorphic DNA and pedigree relationships in spring barley. Theor Appl Genet 85:976–984
Vierling RA, Nguyen HT (1992) Use of RAPD markers to determine the genetic diversity of diploid, wheat genotypes. Theor Appl Genet 84:835–838
Vogel JM, Powell W, Rafalski A, Morgante M, Tudo JD, Taramino G, Biddle P, Hanafey M, Tingley SV (1994) Application of genetic diagnosis to plant genome analysis comparison of marker systems. Appl Biotechnol Tree Cult 1:119–124
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414
Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18:7213–7218
Williams CE, StClair DA (1993) Phenetic relationships and levels of variability detected by restriction fragment length polymorphism and random amplified polymorphic DNA analysis of cultivated and wild accessions of Lycopersicon esculentum. Genome 36:619–630
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Witsenboer H, Kesseli RV, Fortin MG, Stanghellini M, Michelmore RW (1995) Sources and genetic structure of a cluster of genes for resistance to three pathogens in lettuce. Theor Appl Genet 91:1787–188
Yang X, Quiros C (1993) Identification and classification of celery cultivars with RAPD markers. Theor Appl Genet 86:205–212
Zabeau M, Vos P (1993) Selective restriction fragment amplification: a general method for DNA fingerprinting. 924026297 (Publ number 0 534 858 Al)
Author information
Authors and Affiliations
Additional information
Communicated by H.F. Linskens
Rights and permissions
About this article
Cite this article
Hill, M., Witsenboer, H., Zabeau, M. et al. PCR-based fingerprinting using AFLPs as a tool for studying genetic relationships in Lactuca spp.. Theoret. Appl. Genetics 93, 1202–1210 (1996). https://doi.org/10.1007/BF00223451
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00223451