Abstract
Background
Appetite loss has a major impact on cancer patients. It is exceedingly prevalent, is a prognostic indicator and is associated with inferior quality of life. Cachexia is a multi-factorial syndrome defined by a negative protein and energy balance, driven by a variable combination of reduced food intake and abnormal metabolism. Not all cancer patients that experience weight loss have appetite loss, and the pathophysiology between cachexia and appetite loss may thus be different. Knowledge of pathophysiology of appetite loss in cancer patients is still limited. The primary object of this study was to explore the association with 93 predefined candidate single-nucleotide polymorphisms (SNPs) and appetite loss in cancer patients to possibly generate new theories of the pathophysiology of the condition.
Methods
A total of 1,853 cancer patients were phenotyped according to appetite loss and then genotyped.
Results
After allowing for multiple testing, there was no statistically significant association between any of the SNPs analysed and appetite loss. The ten most significant SNPs in the co-dominant model had observed odds ratios varying from 0.72 to 1.28.
Conclusions
This large exploratory study could not find any associations with loss of appetite and 93 SNPs with a potential to be involved in appetite loss in cancer patients. This does not however rule out genes putative role in the development of the symptom, but the observed odds ratios are close to one which makes it unlikely that any of the individual SNPs explored in the present study have great importance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Background
Cachexia has a tremendous impact on survival and morbidity in cancer patients [1]. It is presumed to account for 20 % of cancer deaths due to cardiac and respiratory failure and through immobility [2]. Cachexia is a multi-factorial syndrome defined by a negative protein and energy balance, driven by a variable combination of reduced food intake and abnormal metabolism [3].
Although loss of appetite may occur in some patients, it is by itself not a defining symptom of cachexia. A significant number of cancer patients who lose appetite do not undergo weight loss and quite a few lose weight without having any appetite loss [4, 5]. Consequently, the pathophysiology behind weight loss and appetite loss may be different [6]. Appetite loss is a key symptom in cancer patients: It is exceedingly prevalent [7], is a prognostic indicator [8] and is associated with inferior quality of life [9].
Appetite loss in cancer patients is sometimes divided into primary and secondary anorexia. The pathophysiology behind primary anorexia is still not well understood. It is probably linked to both neurohormonal signalling pathways (e.g. MC4R, serotonin), cytokines [e.g. interleukin-1 (Il-1), tumour necrosis factor alpha (TNF-a)] as well as peripheral appetite signals (e.g. ghrelin, NPY) [10]. Secondary anorexia refers to symptoms that act as barriers to eating [11] for instance nausea, xerostomia, depression and constipation. When studying cancer anorexia and genetic polymorphisms, there are challenges both in phenotyping the patients correctly and in the possibility that both secondary and primary anorexia may coexist.
Cancer cachexia is a very complex condition and thus probably confines both a diverse pathophysiology and phenotype. It has been recognised that the assessment of cachexia should include four domains, namely appetite loss/reduced food intake, catabolic drive, muscle mass and strength as well as functional and psychosocial impairment [3]. By exploring appetite loss as a single factor that contribute to the more complex syndrome of cachexia, one might increase the probability of discovering genotypes and thus the pathopyhysiology implicated in developing the condition. Knowledge of the mechanisms behind cancer anorexia can suggest strategies for prevention, diagnosis and therapy [12] of this widespread and distressing symptom.
There is now growing evidence that appetite and response to food in the general population is partly genetically determined both in obese patients [13, 14] and in patients with psychiatric eating disorders [15]. The impact of genetic polymorphism to the propensity of developing anorexia in cancer patients has only been evaluated in a few studies. Three studies have found associations between appetite loss and TNF-α [16], IL-β [17] and IL-10 [18] gene polymorphisms. In another study, a common mutation in the melanocortin-4 receptor gene (MC4R) was not rendered associated with changes in appetite [19]. None of the polymorphisms found associated with appetite loss in cancer patients have been validated in other cohorts of cancer patients (Table 1).
The primary objective of this study was to explore the association between 93 predefined candidate single-nucleotide polymorphisms (SNPs) and appetite loss in cancer patients to possibly generate new theories of the pathophysiology of the condition.
2 Methods
2.1 Patients and study design
Between February 2004 and April 2008, 2,294 cancer patients were included in the European Pharmacogenetic Opioid Study [20]. This was a multi-centre, cross-sectional, observational study that included patients with cancer at different sites, stages and with different performance status who received opioid treatment. Patients were recruited at 17 centres in 11 different countries. Patients aged <18 years or not capable of understanding the language used at the study centre were not eligible.
The appropriate ethical authorities in all participating centres approved the study protocol, and all patients gave their oral and written informed consent.
2.1.1 Clinical assessment
Age, ethnicity, gender, weight, height, Karnofsky performance status and medication during the last 24 h were recorded. Cancer diagnosis, presence of metastases and time since the diagnosis of cancer were registered. Body mass index was evaluated according to the WHO scale. Information on survival was collected until January 2010. Patients’ subjective health at the time of inclusion was measured by the European Organisation for Research and Treatment of Cancer Core Quality of Life Questionnaire, the EORTC QLQ-C30 version 3.0 [21]. In this questionnaire, the patients report symptoms and functioning for the past week on a four-point verbal rating scale: (1) not at all, (2) a little, (3) quite a bit and (4) very much.
2.1.2 The phenotype for appetite loss
The patients were phenotyped as having anorexia/appetite loss according to the EORTC QLQ-C30 questionnaire (version 3.0) [21, 22], question 13: “have you lacked appetite”.
2.2 Blood sampling
Blood samples were collected at the time of inclusion and stored at −80 °C before shipment to the Norwegian University of Science and Technology (NTNU) in Trondheim, Norway for DNA extraction.
2.2.1 Polymorphisms for exploratory analysis
Prior to the genetic analyses, 93 candidate SNPs from 27 genes were identified through literature review in 2009 as having a putative impact on cancer anorexia pathophysiology. These genes code for appetite regulation through inflammation (TNF, IL-1, Il-6, IL-1, interferon gamma [10], TLR4 [23], hormones (melanocortin, ghrelin, leptin, cholecystokinin, insulin-like growth factor and serotonin [10]) and neuractive receptors (opioid mu receptor [24], cannabinoid receptor [15]). Some SNPs were chosen due to their association with appetite and obesity (peroxisome proliferator-activated receptor gamma, adrenerg receptor [13]) (Online Table 1).
2.2.2 Genotyping and serum analyses
Genomic DNA extraction was performed at HUNT Biobank (NTNU), Levanger, Norway. DNA was extracted from EDTA whole blood using the Gentra Puregene blood kit (Qiagen Science, Germantown, MD, USA).
Genotyping was performed using the SNPlex Genotyping System according to the suppliers’ dry DNA protocol. The capillary electrophoresis was carried out with an ABI 3730 48-capillary DNA analyser. SNPlex signals were analysed using the Gene Mapper version 4.0 software, followed by manual reading. Samples giving low signals, which could not be discriminated from the negative controls, were removed prior to the analysis and treated as missing data. Genotype clustering was performed based on the SNPlex Rules 3730 method, following factory default settings. Two SNPs, rs4680 and rs1045642, which could not be analysed by the SNPlex system, were genotyped at the HUNT Biobank (NTNU), Levanger, Norway. They were analysed using TaqMan SNP allelic discrimination by means of an ABI 7900HT. All genotyping procedures were processed without regard of the phenotype. All genotyping processes was performed according to Applied Biosystems, Foster City, CA, USA
2.3 Statistical analyses
The possible associations with demographic- and disease-related factors were investigated by uni- and multivariate ordinal logistic regressions with appetite loss as the outcome of interest. The factors explored were age, gender, kidney disease, cancer site, Karnofsky status, presence of metastasis and the use of antibiotics, steroids, chemotherapy or appetite stimulants (dronabinol and megestrol) the last 24 h. Factors significantly associated (p < 0.05) with appetite loss in an initial univariate analysis were included in the multivariate analysis. Significant (p < 0.05) factors from the multivariate analyses were included as covariates in the genetic analyses.
The association between SNPs and appetite loss was analysed by ordinal logistic regression with appetite loss as the outcome variable. All regression analysis was stratified by country.
Analyses were also repeated without the inclusion of covariates, as a sensitivity check. Before exploring the genetic associations, SNPs were rejected if the genotypes were not in Hardy–Weinberg equilibrium (HWE) (chi-squared test, p < 0.0005) or had minor allele frequencies (MAF) <5 %.
Three approaches were adopted to mitigate the multiplicity issues. First, a false discovery rate of 10 % was used for reporting the Benjamini–Hochberg (B-H) thresholds [25]. If an SNP is to be classified as “significant”, its p value for association should be below the B-H threshold. Second, the co-dominant genetic model was pre-specified for the primary analyses, with other models (dominant, recessive and additive) being considered as exploratory analyses. A recessive trait will only be expressed if the dominant allele is not present. In a co-dominant model both alleles are visible in the phenotype, and in an additive model, the two alleles have an enhancing effect on each other’s influence on the phenotype. Third, interpretation of p values was done with caution. STATA version 11.0 was used for all analyses (StataCorp. 2009 STATA Statistical Software: Release 11. College Station, TX, USA, StataCorp LP).
3 Results
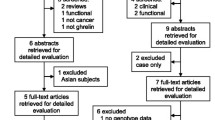
One thousand eight hundred and fifty-three patients with complete data on genetic analyses and appetite score were left for final analyses after excluding five patients from Greece due to their low number, 61 patients of non-caucasian origin 345 patients due to incomplete data on appetite, 24 patients due to missing blood samples and six patients due to no SNPs recorded (Fig. 1). Out of the 1,853 patients left for analyses, 487 patients had no appetite loss, 484 had a little appetite loss, 463 had quite a bit appetite loss and 419 patients had very much appetite loss. Patient demographics are presented in Table 2.
In order to find important covariates, variables potentially associated with appetite loss were found based on clinical experience and literature search. After applying first univariate and then multivariate ordered regression analysis, eight factors were retained as of prognostic importance for grade of appetite loss. These were kidney disease, the use of antibiotics, use of steroids, Karnofsky performance status, cancer originating from female reproductive organs, head and neck cancer, metastasis to liver and metastasis to bone. Age, gender, other metastatic sites, other cancer diagnoses and some medications were not significantly associated and were therefore not included as prognostic factors in the subsequent genetic analyses. Only 282 patients had used chemotherapy the last 24 h and only 28 appetite stimulants; these low numbers may account for the lack of statistical significant impact on appetite, p = 0.055 and p = 0.57 respectively (Table 3).
The 93 candidate SNPs were genotyped in order to identify new associations between appetite loss and genetic polymorphisms. After 12 SNPs were excluded due to violation of HWE or for having a MAF <5 %, 81 candidate SNPs remained for association analysis.
No SNPs were significantly associated with appetite loss neither in the co-dominant, dominant, additive nor recessive model. The ten most significant SNPs in the co-dominant model are presented in Table 4, with observed odds ratios varying from 0.72 to 1.28.
4 Discussion
For the last 10 years, there have been numerous publications on the hereditability of appetite in both a psychiatric eating disorders [15] and obesity [13]. There are only a few studies looking at associations with appetite loss in cancer patients and genetic polymorphisms [16–19]. The present study is the largest study, both in number of patients and number of polymorphisms looking at genetic variability associated with appetite loss in cancer.
In this study, we did not find new associations with appetite loss and polymorphisms with high theoretical potential to be involved in the pathophysiology. The SNP IL-1β-31:rs1143627 that has been found associated with cancer anorexia in a previous study was not significantly associated with appetite loss in the present study [17]
It has, in several studies, been acknowledged that it is notoriously difficult to validate polymorphism associated with common diseases [26] . It is now more strongly recognised that multiple testing and over-reliance on statistical p values results in an excess of false positive associations. This is particularly true when exploratory studies have a small sample size.
If the genetic effect is weak, both a validation study and an explorative study would require very large sample sizes. Thus, even when there is a true association, small-sized studies will overestimate the effect sizes, sometimes grossly so; that is the only way that a significant p value can be achieved in an underpowered study.
If this study had been preformed analysing only a few SNPs, there would probably had been several significant findings. In the present study, we however endeavoured to look at a large sample of patients and have used rigorous statistical methods in order to address the multiplicity issues, the heterogogenecity of different countries, and to take measures for clinical confounding factors. Strong and important relationships between the tested SNPs and appetite loss in cancer patients therefore seem unlikely. There are however numerous SNPs within most genes, and there may still be SNPs within these genes that are important for anorexia.
In genetic studies of common diseases, the capacity to define genotypes is often far better than our capacity to define phenotypes. This is a major concern when conducting and interpreting genetic association studies, and a study looking at appetite is no exception to this. It cannot be ruled out that the lack of association between the SNPs investigated and appetite loss is due to the diagnostic tool used to diagnose appetite loss in the present study. As there is no universal agreement concerning the definition of appetite loss, there are consequently challenges both in defining and diagnosing anorexia. Anorexia and loss of appetite are concepts often intermingled, as both can be defined as the loss of desire to eat [7]. In the present study, we have chosen not to differentiate between anorexia and appetite loss. Anorexia can however also be considered conceptually superior and consist of appetite loss, satiety, taste alterations, altered food preferences [10], reduced food intake [27] and even meat aversions or nausea/vomiting [7]. Appetite loss/anorexia has been endeavoured diagnosed by visual analogue scales and other patient recorded outcomes (e.g. AC/S-12 or FAACT [27]). These assessments all measure slightly different aspects of anorexia and have varying specificity. In addition to this, appetite is often subjectively compared with “normal” for a particular individual, yet there are no thresholds to determine when anorexia becomes present, and there are probably large individual differences. In addition, there is no critical duration of time when to consider the appetite abnormal [10]. These are some of the factors that make it difficult to establish true controls when studying appetite. In the present article, the EORTC-QLQ-C30 question “During the last week - have you lacked appetite?” have been applied to define anorexia. Cancer patients with worse scores on this question have in other studies demonstrated significantly reduced survival [8] and more malnourishment [28]. Another reason for choosing this phenotype is that it has an inherent relation to the symptom in question. The EORTC QLQ-C30 is also a well-validated assessment tool that is translated to many languages.
Secondary anorexia is often defined as symptoms that act as barriers to eating [11] (e.g., nausea, food intolerances and constipation). The causes of secondary anorexia are sometimes the same as the ones suggested contributing to appetite loss. In the present exploratory study, it was therefore chosen not to correct for secondary anorexia in the analysis as one might remove the disease contributing gene (e.g. serotonin is also implicated in depression and nausea [29]; IL-1 may also cause fatigue, constipation and enhance pain [30]; and IL-6 is associated to depression and fatigue [31]).
5 Conclusion
This large exploratory study could not find any associations with loss of appetite and 93 SNPs with a potential to be involved in appetite loss in cancer patients. This does not however rule out genes putative role in the development of the symptom, but the observed odds ratios are close to one which makes it unlikely that any of the individual SNPs explored in the present study have great importance.
In order to move forward in gaining new knowledge on appetite loss in cancer patients, prospective studies with thorough documentation of factors that influence appetite and weight are warranted, as well as consensus on how to diagnose anorexia. This is necessary in order to classify the patients correctly. Unfortunately, there is today no patient-related outcome tool that sufficiently provides this information.
References
Tisdale MJ. Cachexia in cancer patients. Nat Rev Cancer. 2002;2:862–71
Fearon KC. Cancer cachexia: developing multimodal therapy for a multidimensional problem. Eur J Cancer. 2008;44:1124–32.
Fearon K, Strasser F, Anker SD, Bosaeus I, Bruera E, Fainsinger RL et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol. 2011;12:489–495.
Sarhill N, Mahmoud FA, Christie R, Tahir A. Assessment of nutritional status and fluid deficits in advanced cancer. Am J Hosp Palliat Care. 2003;20:465–73.
Bozzetti F. Screening the nutritional status in oncology: a preliminary report on 1,000 outpatients. Support Care Cancer. 2009;17:279–84.
Fordy C, Glover C, Henderson DC, Summerbell C, Wharton R, Allen-Mersh TG. Contribution of diet, tumour volume and patient-related factors to weight loss in patients with colorectal liver metastases. Br J Surg. 1999;86:639–44.
Laviano A, Meguid MM, Rossi-Fanelli F. Cancer anorexia: clinical implications, pathogenesis, and therapeutic strategies. Lancet Oncol. 2003;4:686–94.
Quinten C, Coens C, Mauer M, Comte S, Sprangers MA, Cleeland C, et al. Baseline quality of life as a prognostic indicator of survival: a meta-analysis of individual patient data from EORTC clinical trials. Lancet Oncol. 2009;10:865–71.
Lis CG, Gupta D, Grutsch JF. Can anorexia predict patient satisfaction with quality of life in advanced cancer? Support Care Cancer. 2009;17:129–35.
Davis MP, Dreicer R, Walsh D, Lagman R, LeGrand SB. Appetite and cancer-associated anorexia: a review. J Clin Oncol. 2004;22:1510–7.
Shragge JE, Wismer WV, Olson KL, Baracos VE. The management of anorexia by patients with advanced cancer: a critical review of the literature. Palliat Med. 2006;20:623–9.
Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–8.
Rosado EL, Bressan J, Martins MF, Cecon PR, Martinez JA. Polymorphism in the PPARgamma2 and beta2-adrenergic genes and diet lipid effects on body composition, energy expenditure and eating behavior of obese women. Appetite. 2007;49:635–43.
Tanofsky-Kraff M, Han JC, Anandalingam K, Shomaker LB, Columbo KM, Wolkoff LE, et al. The FTO gene rs9939609 obesity-risk allele and loss of control over eating. Am J Clin Nutr. 2009;90:1483–8.
Helder SG, Collier DA. The genetics of eating disorders. Curr Top Behav Neurosci. 2011;6:157–175.
Jatoi A, Qi Y, Kendall G, Jiang R, McNallan S, Cunningham J, et al. The cancer anorexia/weight loss syndrome: exploring associations with single nucleotide polymorphisms (SNPs) of inflammatory cytokines in patients with non-small cell lung cancer. Support Care Canc. 2009;15:15.
Jatoi A, Nguyen PL, Foster N, Sun D, Stella PJ, Campbell M, et al. Interleukin-1 genetic polymorphisms and their relationship to the cancer anorexia/weight loss syndrome in metastatic gastric and gastroesophageal junction adenocarcinoma. J Support Oncol. 2007;5:41–6.
Rausch SM, Clark MM, Patten C, Liu H, Felten S, Li Y et al. Relationship between cytokine gene single nucleotide polymorphisms and symptom burden and quality of life in lung cancer survivors. Cancer. 2010;116:4103–4113.
Knoll S, Zimmer S, Hinney A, Scherag A, Neubauer A, Hebebrand J. Val103Ile polymorphism of the melanocortin-4 receptor gene (MC4R) in cancer cachexia. BMC Cancer. 2008;8:85.
Klepstad P FT, Skorpen F, Bjordal K, Caraceni A, Dale O, Davies A, Kloke M, Lundström S, Maltoni M, Radbruch L, Sabatowski R, Sigurdardottir V, Strasser F, Fayers PM, Kaasa S on behalf of the European Palliative Care Research Collaborative (EPCRC) and the European Association for Palliative Care Research Network. The influence from genetic variability on opioid use for cancer pain. An European study of 2294 cancer pain patients. Pain. 2011;152:1139–1145.
Aaronson NK, Ahmedzai S, Bergman B. The European organization for research and treatment of cancer QLQ-C30: a quality-of-life instrument for use in international clinical trials in oncology. J Natl Canc Inst. 1993;85:365–76.
Fayers P AN, Bjordal K, Sullivan M. EORTC QLQ-C30 scoring manual. EORTC Study Group on Quality of Life, Brussels. 1995;1–49.
Langhans W. Signals generating anorexia during acute illness. Proc Nutr Soc. 2007;66:321–30.
Gosnell BA, Levine AS. Reward systems and food intake: role of opioids. Int J Obes (Lond). 2009;33(Suppl 2):S54–S58.
Benjamini YHY. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300.
Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61.
Muscaritoli M, Anker SD, Argiles J, Aversa Z, Bauer JM, Biolo G et al. Consensus definition of sarcopenia, cachexia and pre-cachexia: Joint document elaborated by Special Interest Groups (SIG) “cachexia–anorexia in chronic wasting diseases” and “nutrition in geriatrics”. Clin Nutr. 2010;29:154–159.
Capuano G, Gentile PC, Bianciardi F, Tosti M, Palladino A, Di Palma M. Prevalence and influence of malnutrition on quality of life and performance status in patients with locally advanced head and neck cancer before treatment. Support Care Canc. 2009;18:433–7.
Ueda N, Yoshimura R, Shinkai K, Sakata Y, Nakamura J. Higher plasma 5-hydroxyindoleacetic acid levels are associated with SSRI-induced nausea. Neuropsychobiology. 2003;48:31–4.
Seruga B, Zhang H, Bernstein LJ, Tannock IF. Cytokines and their relationship to the symptoms and outcome of cancer. Nat Rev Cancer. 2008;8:887–99.
Valentine RJ, Woods JA, McAuley E, Dantzer R, Evans EM. The associations of adiposity, physical activity and inflammation with fatigue in older adults. Brain Behav Immun. 2011;25:1482–1490.
von Haehling SMJ, Coats AJ, Anker SD. Ethical guidelines for authorship and publishing in the Journal of Cachexia, Sarcopenia and Muscle. J Cachexia Sarcopenia Muscle. 2010;1:7–8.
Acknowledgments
The authors thank Daniel L. Marks, Oregon, USA and William F Colmers, Alberta, Canada for valuable discussions concerning candidate SNPs. The authors of this manuscript certify that they comply with the ethical guidelines for authorship and publishing in the Journal of Cachexia, Sarcopenia and Muscle [32].
Conflicts of interest
The authors declare that they have no conflict of interest
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Please see Appendix 1 for details of the European Palliative Care Research Collaborative (EPCRC) and the European Pharmacogenetic Study (EPOS).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online table 1
(DOC 233 kb)
Appendix 1. Research collaboratives
Appendix 1. Research collaboratives
1.1 European Pharmacogenetic Opioid Study
Principal investigator
Pål Klepstad, Trondheim, Norway.
Investigator at each centre
Andrew Davies, Sutton/Chelsea, UK; Augusto Caraceni, Alessandra Pigni, Milan, Italy; Danilo Miotti, Pavia, Italy; Eeva Salminen, Turku, Finland; Eriphili Argyra, Athens, Greece; Florian Strasser, St. Gallen Switzerland; Irena Poviloniene, Vilnius, Lithuania; Jon Håvard Loge, Oslo, Norway; Kristin Bjordal, Oslo, Norway; Lucas Radbruch, Aachen, Germany; Marianne Kloke, Essen Germany; Marco Maltoni, Forli, Italy; Per Sjøgren, Copenhagen Denmark; Rainer Sabatowski, Dresden, Germany; Staffan Lundström, Stockholm, Sweden; Stein Kaasa, Trondheim, Norway; Valgerdur Sigurdardottir, Reykjavik, Iceland.
1.2 European Palliative Care Research Collaborative
The European Palliative Care Research Collaborative is funded by the European Commission’s Sixth Framework Programme (contract no LSHC-CT-2006-037777) with the overall aim to improve treatment of pain, depression and fatigue through translation research.
Core scientific group/work package leaders
Stein Kaasa (project coordinator), Frank Skorpen, Marianne Jensen Hjermstad, and Jon Håvard Loge, Norwegian University of Science and Technology (NTNU); Geoffrey Hanks, University of Bristol; Augusto Caraceni and Franco De Conno, Fondazione IRCCS Istituto Nazionale dei Tumori, Milan; Irene Higginson, King’s College London; Florian Strasser, Cantonal Hospital St. Gallen; Lukas Radbruch, RWTH Aachen University; Kenneth Fearon, University of Edinburgh; Hellmut Samonigg, Medical University of Graz; Ketil Bø, Trollhetta AS, Norway; Irene Rech-Weichselbraun, Bender MedSystems GmbH, Austria; Odd Erik Gundersen, Verdande Technology AS, Norway.
Scientific advisory group
Neil Aaronson, The Netherlands Cancer Institute; Vickie Baracos and Robin Fainsinger, University of Alberta; Patrick C. Stone, St. George’s University of London; Mari Lloyd-Williams, University of Liverpool.
Project management
Stein Kaasa, Ola Dale, and Dagny F. Haugen, NTNU.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Solheim, T.S., Fayers, P.M., Fladvad, T. et al. Is there a genetic cause of appetite loss?—an explorative study in 1,853 cancer patients. J Cachexia Sarcopenia Muscle 3, 191–198 (2012). https://doi.org/10.1007/s13539-012-0064-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13539-012-0064-8