Abstract
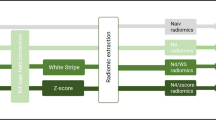
Glioblastoma multiforme (GBM) is the most lethal and aggressive brain tumor. Magnetic resonance imaging (MRI) is currently used to diagnose and monitoring it. Radiomic features extracted from MRI are being used for correlations with the disease prognosis. A methodology to extract MRI radiomics features in a radiotherapy planning workflow, to assess features related to GBM poor prognosis patients, is presented. One hundred five radiomics features were extracted from T1 post-contrast MRIs of 43 GBM patients. The progression-free survival (PFS) time of all patients was also achieved. These patients were separated into two groups: PFS within 3 months (class value = 1) and PFS higher than 3 months (class value = 0). A machine learning model was built to predict the poor prognosis of patients using a random forest algorithm optimized for class = 1 classification, i. e. optimized for the recall score. Kurtosis was ranked as the most important feature for this classification. The final model presents a recall score of 1.00 ± 0.0 and an area under a receiver operating characteristic curve (AUROC) of 0.81 ± 0.04. The model used a kurtosis threshold value at 2.69 ± 0.03 for classification. The kurtosis values achieved for the PFS within 3 months indicate that these tumors are composed of almost the same amount of necrose, neoangiogenesis, edema, and/or tumor cells. In conclusion, a relation between the radiomics analysis of kurtosis in MRI with GBM’s poor prognosis was found, and the developed model may help guide the patient’s clinical management.





Similar content being viewed by others
References
A. Chaddad, M.J. Kucharczyk, P. Daniel, S. Sabri, B.J. Jean-Claude, T. Niazi et al., Radiomics in glioblastoma: current status and challenges facing clinical implementation. Front. Oncol. 9, 1–9 (2019). https://doi.org/10.3389/fonc.2019.00374
R. Stupp, M.E. Hegi, W.P. Mason, M.J. van den Bent, M.J. Taphoorn, R.C. Janzer et al., Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10, 459–66 (2009). https://doi.org/10.1016/S1470-2045(09)70025-7
C. Adamson, O.O. Kanu, A.I. Mehta, C. Di, N. Lin, A.K. Mattox et al., Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin. Investig. Drugs. 18, 1061–83 (2009). https://doi.org/10.1517/13543780903052764
A. Sottoriva, I. Spiteri, S.G.M. Piccirillo, A. Touloumis, V.P. Collins, J.C. Marioni et al., Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. 110, 4009–14 (2013). https://doi.org/10.1073/pnas.1219747110
S. Moton, M. Elbanan, P.O. Zinn, R.R. Colen, Imaging genomics of glioblastoma. Top. Magn. Reson. Imaging. 24, 155–63 (2015). https://doi.org/10.1097/RMR.0000000000000052
R.J. Gillies, A.R. Anderson, R.A. Gatenby, D.L. Morse, The biology underlying molecular imaging in oncology: from genome to anatome and back again. Clin. Radiol. 65, 517–21 (2010). https://doi.org/10.1016/j.crad.2010.04.005
H.J.W.L. Aerts, E.R. Velazquez, R.T.H. Leijenaar, C. Parmar, P. Grossmann, S. Cavalho et al., Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, (2014). https://doi.org/10.1038/ncomms5006
P. Lambin, R.G.P.M. Van Stiphout, M.H.W. Starmans, E. Rios-Velazquez, G. Nalbantov, H.J.W.L. Aerts et al., Predicting outcomes in radiation oncology-multifactorial decision support systems. Nat. Rev. Clin. Oncol. 10, 27–40 (2013). https://doi.org/10.1038/nrclinonc.2012.196
H. Sotoudeh, O. Shafaat, J.D. Bernstock, M.D. Brooks, G.A. Elsayed, J.A. Chen et al., Artificial intelligence in the management of glioma: era of personalized medicine. Front. Oncol. 9, (2019). https://doi.org/10.3389/fonc.2019.00768
C.A. Sarkiss, I.M. Germano, Machine learning in neuro-oncology: can data analysis from 5346 patients change decision-making paradigms? World Neurosurg. 124, 287–94 (2019). https://doi.org/10.1016/j.wneu.2019.01.046
D. Assefa, H. Keller, C. Ḿnard, N. Laperriere, R.J. Ferrari, I. Yeung, Robust texture features for response monitoring of glioblastoma multiforme on T1 -weighted and T2 -FLAIR MR images: a preliminary investigation in terms of identification and segmentation. Med. Phys. 37, 1722–36 (2010). https://doi.org/10.1118/1.3357289
S. Bauer, T. Fejes, J. Slotboom, R. Wiest, L.-P. Nolte, M. Reyes, Segmentation of brain tumor images based on integrated hierarchical classification and regularization. MICCAI BraTS Work. 10–3 (2012)
K. Li-Chun Hsieh, C.-Y. Chen, C.-M. Lo, Quantitative glioma grading using transformed gray-scale invariant textures of MRI. Comput. Biol. Med. 83, 102–8 (2017). https://doi.org/10.1016/j.compbiomed.2017.02.012
J. Wu, Z. Qian, L. Tao, J. Yin, S. Ding, Y. Zhang et al., Resting state fMRI feature-based cerebral glioma grading by support vector machine. Int. J. Comput. Assist. Radiol. Surg. 10, 1167–74 (2015). https://doi.org/10.1007/s11548-014-1111-z
Q. Tian, L.-F. Yan, X. Zhang, X. Zhang, Y.-C. Hu, Y. Han et al., Radiomics strategy for glioma grading using texture features from multiparametric MRI. J. Magn. Reson. Imaging. 48, 1518–28 (2018). https://doi.org/10.1002/jmri.26010
P.A. Eliat, D. Olivié, S. Saïkali, B. Carsin, H. Saint-Jalmes, J.D. De Certaines, Can dynamic contrast-enhanced magnetic resonance imaging combined with texture analysis differentiate malignant glioneuronal tumors from other glioblastoma? Neurol. Res. Int. (2012). https://doi.org/10.1155/2012/195176
E.I. Zacharaki, S. Wang, S. Chawla, D.S. Yoo, R. Wolf, E.R. Melhem et al., Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. 62, 1609–18 (2009). https://doi.org/10.1002/mrm.22147
M. Zhou, L. Hall, D. Goldgof, R. Russo, Y. Balagurunathan, R. Gillies et al., Radiologically defined ecological dynamics and clinical outcomes in glioblastoma multiforme: Preliminary results. Transl. Oncol. 7, 5–13 (2014). https://doi.org/10.1593/tlo.13730
A. Chaddad, S. Sabri, T. Niazi, B. Abdulkarim, Prediction of survival with multi-scale radiomic analysis in glioblastoma patients. Med. Biol. Eng. Comput. 56, 2287–300 (2018). https://doi.org/10.1007/s11517-018-1858-4
S.S.F. Yip, H.J.W.L. Aerts, Applications and limitations of radiomics. Phys. Med. Biol. 61, R150-66 (2016). https://doi.org/10.1088/0031-9155/61/13/R150
J.J.M. van Griethuysen, A. Fedorov, C. Parmar, A. Hosny, N. Aucoin, V. Narayan et al., Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77, e104-7 (2017). https://doi.org/10.1158/0008-5472.CAN-17-0339
S.S. Shapiro, M.B. Wilk, An analysis of variance test for normality (complete samples). Biometrika. 52, 591 (1965). https://doi.org/10.2307/2333709
F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel et al., Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–30 (2011)
C.H.B. Liu, B.P. Chamberlain, D.A. Little, Â. Cardoso, Generalising random forest parameter optimisation to include stability and cost. p. 102–13 (2017). https://doi.org/10.1007/978-3-319-71273-4_9
M. Wehenkel, A. Sutera, C. Bastin, P. Geurts, C. Phillips, Random forests based group importance scores and their statistical interpretation: application for Alzheimer’s disease. Front. Neurosci. 12, (2018). https://doi.org/10.3389/fnins.2018.00411
G. James, D. Witten, T. Hastie, R. Tibshirani, An Introduction to statistical learning. vol. 103. New York, NY: Springer New York. (2013). https://doi.org/10.1007/978-1-4614-7138-7
H. He, Y. Ma, editors. Imbalanced Learning. Wiley. (2013). https://doi.org/10.1002/9781118646106
S. Bae, Y.S. Choi, S.S. Ahn, J.H. Chang, S.G. Kang, E.H. Kim et al., Radiomic MRI phenotyping of glioblastoma: improving survival prediction. Radiology 289, 797–806 (2018). https://doi.org/10.1148/radiol.2018180200
X. Chen, M. Fang, D. Dong, L. Liu, X. Xu, X. Wei et al., Development and validation of a MRI-based radiomics prognostic classifier in patients with primary glioblastoma multiforme. Acad. Radiol. 26, 1292–300 (2019). https://doi.org/10.1016/j.acra.2018.12.016
M.W. McDonald, H.-K.G. Shu, W.J. Curran, I.R. Crocker, Pattern of failure after limited margin radiotherapy and temozolomide for glioblastoma. Int. J. Radiat. Oncol. 79, 130–6 (2011). https://doi.org/10.1016/j.ijrobp.2009.10.048
B.J. Gebhardt, M.C. Dobelbower, W.H. Ennis, A.K. Bag, J.M. Markert, J.B. Fiveash, Patterns of failure for glioblastoma multiforme following limited-margin radiation and concurrent temozolomide. Radiat. Oncol. 9, 130 (2014). https://doi.org/10.1186/1748-717X-9-130
M.C. Dobelbower, O.L. Burnett III., R.A. Nordal, L.B. Nabors, J.M. Markert, M.D. Hyatt et al., Patterns of failure for glioblastoma multiforme following concurrent radiation and temozolomide. J. Med. Imaging Radiat. Oncol. 55, 77–81 (2011). https://doi.org/10.1111/j.1754-9485.2010.02232.x
H. Um, F. Tixier, D. Bermudez, J.O. Deasy, R.J. Young, H. Veeraraghavan, Impact of image preprocessing on the scanner dependence of multi-parametric MRI radiomic features and covariate shift in multi-institutional glioblastoma datasets. Phys. Med. Biol. 64, (2019). https://doi.org/10.1088/1361-6560/ab2f44
S. Narang, M. Lehrer, D. Yang, J. Lee, A. Rao, Radiomics in glioblastoma: current status, challenges and potential opportunities. Transl. Cancer Res. 5, 383–97 (2016). https://doi.org/10.21037/tcr.2016.06.31
J. Pyyry, W. Keranen, Varian APIs. First Edit. (2018)
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
de Marco Borges, P.H., Lizar, J.C., Faustino, A.C.C. et al. Kurtosis is An MRI Radiomics Feature Predictor of Poor Prognosis in Patients with GBM. Braz J Phys 51, 1035–1042 (2021). https://doi.org/10.1007/s13538-021-00912-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13538-021-00912-9