Abstract
Background
The role of Notch signaling in HPV-mediated transformation has been a long standing debate, as both tumor suppressive and oncogenic properties have been described. We examined whether the dual findings in literature may be explained by gene dosage effects and determined the relation with AP-1, a downstream target of Notch.
Methods
SiHa cervical cancer cells were transfected with two doses of intracellular active Notch. Non-tumorigenic HPV16-immortalized keratinocytes (FK16A) were transfected with Fra1 specific siRNAs and non-targeting controls. Transfectants were analysed for Notch, Hes, cJun, cFos and Fra1 mRNA expression, Notch pathway activation using luciferase assays, cell viability using MTT assays, anchorage independent growth, AP-1 activity and/or AP-1 complex composition using EMSA.
Results
In SiHa cells two activation states of Notch signaling pathway were obtained. Moderate Notch activation contributed to increased viability and anchorage independent growth, whereas high level Notch activation decreased anchorage independent growth. The shift in phenotypical outcome was correlated to altered AP-1 activity and complex composition. Moderate Notch expression led to an increased AP-1 transcriptional activity and DNA binding activity, but did not affect complex composition. High levels of Notch additionally led to a change in AP-1 complex composition, from cJun/cFos to cJun/Fra1 dimers, which is exemplary for non-tumorigenic HPV-immortalized cell lines. Conversely, silencing of Fra1 in non-tumorigenic HPV16-immortalized keratinocytes, leading to an enrichment of cJun/cFos dimers, was accompanied with increased colony formation.
Conclusion
The functional role of Notch in HPV-mediated transformation is dosage dependent and correlated to a change in AP-1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Sustained expression of the human papilloma virus (HPV) oncogenes E6 and E7 is necessary to acquire and maintain the transformed state of cervical cancer and its high grade precursor lesions. However, additional cellular alterations are required for the progression of HPV-containing precursor lesions towards cervical cancer. Several studies indicate the involvement of aberrant Notch signaling in the process of cervical carcinogenesis [3, 6, 10, 15, 16, 18, 19, 21]. Notch plays a role in multiple cellular processes, such as differentiation, proliferation and apoptosis (reviewed by [8]). Upon ligand interaction of Delta and Jagged/Serrate family members, the transmembrane receptor Notch is conformationally changed which allows proteolytic cleavage and release of the intracellular part of Notch (ICNX). After translocation to the nucleus, intracellular Notch competes with repressor proteins for binding the transcription factor C-promoting binding factor 1 (CBF1). Binding of ICNX to CBF1 leads to recruitment of coactivators which turn CBF1 from a transcriptional repressor into an activator, mainly targeting the transcription family Hairy enhancer of split (Hes).
In cervical cancer, both tumor suppressive as well as oncogenic properties of Notch signaling have been described. Some studies report undetectable Notch levels in cervical cancer cell lines and reduced expression in cancer specimens. In these studies Notch overexpression has tumor suppressive effects, such as growth inhibition and induction of apoptosis [15, 16, 18]. Others have demonstrated that increased Notch expression correlates with progression to cervical cancer [3, 9, 21] and contributes to transformation by signaling for survival [10, 19]. At least part of the discrepancies in functional studies may be explained by dosage effects, as it has been previously shown that high Notch levels reduced proliferation and HPV promoter activity in cervical cancer cells, whereas moderate Notch expression levels enhanced transformation of HPV transfected keratinocytes [6].
One of the downstream effectors of Notch is the AP-1 transcription factor complex. This is a dimeric complex consisting of both Jun family members (cJun, JunD, JunB) and Fos family members (Fra1, Fra2, cFos, FosB) [5]. The AP-1 complex can bind to the TPA response element (TRE), thereby regulating gene transcription. Genes containing such elements are involved in various cellular processes such as differentiation, proliferation and apoptosis. The effect of AP-1 on gene expression depends on the dimerization partners, the promoter and other transcription factors involved, as well as the cell type analysed. AP-1 is also involved in transcriptional regulation of HPV, as the upstream regulatory region (URR) of HPV harbors AP-1 binding sites in the promoter and enhancer region [11, 17]. Moreover, AP-1 complex composition appears phenotype dependent in HPV-containing cells. Whereas tumorigenic cervical cancer cells like SiHa and HeLa were found to contain cJun/cFos dimers, non tumorigenic HeLa-fibroblast hybrids as well as non-tumorigenic HPV-immortalized keratinocytes displayed cJun/Fra1 dimers [11, 12, 20].
In previous work, we demonstrated that an altered AP-1 composition seen between non-tumorigenic HPV-immortalized keratinocytes and tumorigenic cervical cancer cells was associated with a change in Notch mRNA expression [20]. Notch expression levels progressively increased with hrHPV-mediated immortalization of primary keratinocytes in vitro. However, Notch expression was strongly reduced in cervical cancer cell lines SiHa and HeLa [20]. The fact that Notch expression increases during early stages of transformation but is reduced in cancer cell lines, suggests that its functional effects may be dose-dependent. In the present study we aimed to investigate the dosage dependent functional effects of Notch in HPV-containing cells and to determine the relation to AP-1.
2 Material and methods
2.1 Cell culture, transfection and luciferase assays
Cervical cancer cell line SiHa was obtained from the American Type Culture Collection (Manasas, VA, USA) and cultured in DMEM supplemented with 10% fetal calf serum, penicillin (100U/ml), streptomycin (100 μg/ml), and L-glutamine (2 mM). HPV16-immortalized keratinocytes (FK16A) were cultured in serum-free keratinocyte medium (Invitrogen, Breda, The Netherlands), as described previously [14]. Transfections were performed using Lipofectamine (Invitrogen) or Dharmafect2 (Dharmacon, Lafayette, Louisiana, USA) for cDNA and siRNA respectively, according to the manufacturers instructions. cDNA encoding the intracellular part of Notch was a kind gift from dr. Warren Pear (Department of Pathology, University of Pennsylvania, Philadelphia, USA) and was cloned into pcDNA3.1hygro expression vector (Invitrogen). Moderate Notch expression levels were obtained by transfecting according to manufacturers instructions (4 μg DNA/2500 μl medium/6 well). High expression levels of Notch were created by increasing the concentration of transfection mixture 5 times (4 μg DNA/500 μl medium/6 well). Empty vector was used in both concentrations as a control. A pool of Fra-1 specific siRNAs was used to silence Fra1 (#L-004341-00-0005, Dharmacon). A pool of 4 non-targeting siRNAs (#D-001810-10-05, Dharmacon) was used as negative control. The use of a non-targeting siRNA pool ensures control for off-target effects. The luciferase construct for Hes1 was kindly provided by dr. Marc Vooijs (Department of Pathology, University Medical Center Utrecht, The Netherlands) and URR-luciferase was obtained from Dr. Felix Hoppe-Seyler (Department of Molecular Therapy of Virus-Associated Cancers, German Cancer Research Center, Heidelberg, Germany). Firefly luciferase and Renilla luciferase were measured using Dual Luciferase assay (Promega, Wisconsin, USA).
2.2 RNA isolation and RT-PCR
mRNA was isolated using RNA-B reagent (Tel-Test, Friendswood, TX, USA). Isolated RNA was DNase treated using RQ1 RNase-free DNase (Promega, Madison, WI, USA) and used for cDNA synthesis with a specific reverse primer. Quantitative RT-PCR for AP-1 family members was performed as described previously using primers and probes as shown in Table 1 [20]. To correct for RNA quality and input, we performed RT-PCR for the housekeeping gene snRNP U1A. For quantification purposes a standard curve was made of serial dilutions of cell line cDNA of cell lines highly expressing the target (primary keratinocytes, FK16A or SiHa) synthesized with the respective specific reverse primers. Notch and Hes1 RT-PCRs were performed using SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA) and AP-1 member RT-PCRs were performed using Universal PCR Master Mix (Applied Biosystems).
2.3 EMSA
Nuclear extracts were prepared as described previously [2]. Protein concentration was determined using the BCA protein assay kit from Pierce (Thermo Fisher Scientific, Rockford, IL, USA). AP-1 consensus oligonucleotide was generated in an Applied Biosystems synthesizer using phosphoramitide chemistry and further purified by HPLC. The annealed oligonucleotide was labeled with [γ-32P]ATP (Amersham, 3000 Ci/mmol) using T4 polynucleotide kinase and purified from a 15% polyacrylamide gel. The binding reaction was performed as described before [4]. After 30 min incubation at room temperature, 2 μg of specific antibody was added and incubated at 4°C for 1 h. The antibody used for cJun was sc822x, for Fra1 sc605x and for cFos sc52x (all from Santa Cruz Biotechnology Inc, Santa Cruz, USA). The complexes were separated on 5.5% nondenaturing polyacrylamide gels by running 30 min at 280 V and 75 min at 350 V.
2.4 Viability and anchorage independent growth
Cell viability was assessed by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) dye reduction (ICN Biomedicals Inc, OH, USA). Cells were seeded in triplicate wells in 96-wells plates after transfection and measured after 5 days in culture. Control conditions were set to 100%.
To examine anchorage independent growth under the different conditions 5000 cells were plated in semi-solid agarose as described before [13]. After 3 weeks colonies exceeding ~50 cells were counted and pictures taken.
2.5 Statistical analysis
All statistical analyses were carried out using the T-test in Excel. A p-value ≤0.05 was considered statistically significant.
3 Results
3.1 Dose-dependent effects of ectopic Notch expression in SiHa cervical cancer cells
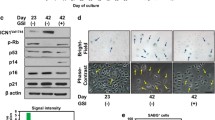
To analyze the potential dose dependent effects of Notch in HPV-induced transformation, moderate and high levels of ectopic Notch expression were examined in the cervical cancer cell line SiHa, which is known to have reduced Notch levels [20]. Towards this end two different concentrations of cDNA encoding the intracellular active form of Notch (ICNX) were transfected in SiHa cells, referred to as moderate and high. Moderate ICNX concentrations resulted in 46 times increased Notch mRNA expression and transfection of high concentrations of ICNX resulted in 191 times increased Notch mRNA expression (Fig. 1a).
Dose-dependent effect of Notch expression in SiHa cells. a Notch mRNA expression levels upon transfection of two concentrations of transfection mix. ev: empty vector control b Relative luciferase activity of Hes1 responsive luciferase reporter plasmid in SiHa cells transfected with two doses of Notch compared to untransfected (untf) cells. c Messenger RNA expression of the downstream target of Notch signaling, Hes1. ev: empty vector control
To next examine the dose-dependent effect of Notch overexpression on pathway activation, co-transfections of ICNX and luciferase constructs driven by the Hes1 promoter were performed in SiHa cells. Figure 1b shows that Notch induced Hes1 transcription in a dose-dependent manner from 15-fold to 34-fold compared to untransfected controls in cells with moderate and high Notch, respectively. Also Hes1 mRNA expression was increased in Notch transfectants, confirming functional Notch activity. However, no difference in Hes1 mRNA levels was observed between cells with moderate and high Notch (Fig. 1c), suggesting that regulatory mechanisms other than those measured by reporter assays, such as chromatin and post-transcriptional modifications, may refrain Hes1 mRNA expression from being further upregulated.
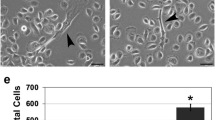
To determine the functional consequences of ectopic Notch expression SiHa cells transfected with the two ICNX concentrations and corresponding empty vector concentrations were assayed for cellular viability using an MTT assay and anchorage independent growth. Moderate Notch expression resulted in a significantly higher cell viability compared to empty vector control cells (p = 0.0002). Conversely, high Notch expression did not alter cell viability (Fig. 2a). Colony formation was altered in different ways at both conditions; moderate levels of Notch increased the colony forming capacity (not significant), whereas high expression levels of Notch resulted in a significantly reduced colony forming capacity (p = 0.05) (Fig. 2b).
Functional effects of moderate and high Notch overexpression in SiHa cells. a Cellular viability measured by MTT assay. The Y-axis indicates the relative cellular viability following 5 days in culture. Control transfections were set to 100% b Quantification of anchorage independent growth, in which the number of colonies was counted after 3 weeks in culture. Control transfections were set to 100%. (*) Indicates significant differences compared to control transfections
3.2 Effects of Notch on AP-1 transcription, activity and composition are dose-dependent
Next, we determined transcriptional activity of AP-1, a well known downstream target of Notch signaling, upon Notch overexpression. Co-transfection experiments of ICNX cDNA and a TPA response element (TRE) driven luciferase construct, the latter allowing to measure AP-1 activity, revealed that AP-1 activity was induced in a dose-dependent manner (Fig. 3a).
Effect of moderate and high Notch expression on AP-1 activity, AP-1 member mRNA expression and complex composition. a Relative luciferase activity of a TPA-response element (TRE) - responsive luciferase reporter plasmid in SiHa cells transfected with moderate and high levels of Notch compared to untransfected (untf) controls (set to 1). b mRNA expression of AP-1 family members cJun, Fra1 and cFos upon moderate and high Notch expression compared to cells transfected with empty vector (set to 1). c EMSA performed using nuclear extracts of SiHa transfected with empty vector DNA, moderate and high levels of Notch cDNA. Specific antibodies were used to obtain supershifts for the different components
Whereas AP-1 can bind to TPA response elements also transcription of AP-1 members itself is regulated by these TREs. To determine whether increasing levels of Notch expression affected mRNA expression of AP-1 family members, quantitative RT-PCR for different family members was performed (Fig. 3b). Induction of moderate Notch overexpression resulted in increased mRNA expression of cJun, while cFos and Fra1 mRNA expression were not markedly altered. Upon high Notch expression, cJun mRNA was still increased, but to a lower level compared to moderate Notch expression. Also a slight increase in cFos mRNA was seen, whereas Fra1 mRNA expression remained unchanged.
To determine whether Notch overexpression was correlated to altered AP-1 DNA binding activity and complex composition Electrophoretic Mobility Shift Assays (EMSAs) were performed. As shown in the empty vector control transfectants (Fig. 3c, left panel), the AP-1 complexes in SiHa cells consist of cJun/Fra1, cJun/cFos and possibly also cJun/cJun dimers. After inducing a moderate level of Notch expression, an increased DNA binding was observed (Fig. 3c, compare lane 5 with lane 1), indicating elevated AP-1 activity, in line with the luciferase assays (Fig. 3a). However, no change in composition was seen. Strikingly, cells with high expression levels of Notch displayed no visible increase in DNA binding activity though the cFos signal was decreased and the Fra1 signal was increased, indicating a shift in complex composition towards cJun/Fra1. This change is likely to reflect a functionally altered AP-1 complex as an increase in cJun/cFos dimers in the AP-1 complex is associated with a tumorigenic phenotype, while a complex primarily consisting of cJun/Fra1 dimers is linked to a non-tumorigenic state [11, 20].
3.3 Silencing Fra1 in HPV16-immortalized keratinocytes alters AP-1 and increases anchorage independent growth
Human foreskin keratinocytes immortalized by full length HPV16 posses an AP-1 complex consisting of cJun/Fra1 and are non-tumorigenic [20]. To examine the functional consequence of altered AP-1 complexes in HPV16-immortalized cells we depleted Fra1 in immortal FK16A cells (passage 156) by transfecting a pool of siRNAs targeting Fra1 or non-targeting siRNAs as control.
Fra1 silencing resulted in an upregulation of cFos mRNA and had little effect on cJun mRNA expression (Fig. 4a). Examination of AP-1 complex composition in these cells showed that silencing of Fra1 resulted in a reduced Fra1 incorporation into the complex, with more cJun/cFos dimers being the consequence (Fig. 4b). HPV16-immortalized keratinocytes silenced for Fra1 displayed increased colony forming capacity in soft agarose (Fig. 4c) without an increase in proliferative potential (not shown). No effects were seen using non-targeting siRNAs on complex composition, proliferation or anchorage independent growth. These results further strengthen the notion that changes in AP-1 complex composition are important in HPV-mediated transformation.
The effect of Fra1 silencing in HPV16-immortalized keratinocytes. a mRNA expression of AP-1 family members Fra1, cFos and cJun upon transfection with siRNAs targeting Fra1 or a non-targeting siRNA control. b EMSA for AP-1 composition using cells transfected with siRNAs targeting Fra1 compared to non-targeting control. The lower band is total AP-1 bound to the consensus oligo. Arrows indicate specific antibody supershifts. c Colony formation in soft agarose. Transfection with siRNAs targeting PLK1 which induces cell death was included as a control for transfection efficiency
4 Discussion
The dual findings on the role of Notch signaling in cervical carcinogenesis have been a long standing debate. Recently, a model has been proposed by Maliekal et al., suggesting that the opposing effects described in literature are dosage dependent [7]. This model was based on the application of a single dose of Notch expression in individual cell lines with different outcomes in the different studies [15, 16, 18, 19] and on the effects of two levels of Notch overexpression in HeLa cells and HaCat cells, respectively, on DNA synthesis and HPV promoter activity [6]. However, there has been no direct proof for this concept. Here, we analysed this hypothesis directly by studying the effects of two Notch expression levels on several phenotypes in the same cell line. Two expression levels of intracellular active Notch were created leading to two activation states of the signaling pathway, as confirmed using Hes luciferase assays, in the cervical cancer cell line SiHa. While moderate Notch activation contributed to increased viability and anchorage independent growth properties, high level Notch activation exhibited tumor suppressive traits, e.g. reduced colony formation. Hence, these findings nicely confirm the model as proposed by Maliekal et al., and provide at least in part an explanation for the controversial data published previously. Moreover, we showed that the dual findings in phenotypical outcome upon moderate and high Notch expression may be explained by a switch in composition of the transcription factor AP-1, a well known downstream target of Notch signaling. In tumorigenic SiHa cells, cJun/cFos heterodimers are more prominent than in non-tumorigenic HPV-immortalized cell lines. Increasing Notch activation to moderate levels in these cells increased AP-1 activity, as measured using TRE-driven luciferase, and DNA binding, as seen in EMSAs, though did not affect complex composition. High level Notch activation, on the other hand, led to a change in the composition of AP-1, towards cJun/Fra1.
The correlation between Notch and AP-1 family member expression and function has been the subject of several studies, though has not yet been analysed in the context of AP-1 complex composition. It has been demonstrated that Notch overexpression in HeLa cells resulted in increased protein expression of cJun [18], which is in line with the presently observed increase in mRNA expression upon moderate Notch expression in SiHa cells. However, Notch overexpression was previously found to decrease the expression of cFos in HeLa cells [15, 16, 18], and linked to decreased AP-1 activity [18]. This is in contrast to present findings in which AP-1 activity was increased upon both moderate and high Notch expression, whereas also c-Fos mRNA was slightly increased. On the other hand, in the study by Talora et al., AP-1 activity was not affected upon Notch overexpression in SiHa, HeLa and CaSki cells [15].Therefore, the absence of concordance between the various studies on Notch and AP-1 activity, indicates that neither expression of individual AP-1 members, nor sole AP-1 activity measured by reporter-assays can explain the link between AP-1 and Notch in HPV-mediated transformation, and that also complex composition should be taken into account.
The importance of complex composition with respect to phentoypical outcome has been demonstrated in HPV16-immortalized keratinocytes in which cJun/Fra1 complexes are prominent [20]. Silencing of Fra1 using siRNAs reduced AP-1 DNA binding activity and reduced Fra1 incorporation in EMSA, while colony formation was increased. The resulting enrichment of cJun/cFos dimers led to increased anchorage independent growth properties. These results are in line with those described by Soto et al., for HeLa cells [12].
Part of the discrepancies in literature may also be related to the cell lines used. CaSki cells express much higher levels of both Notch and Fra1 compared to SiHa and HeLa cells [20]. Notch expression appeared essential for the tumorigenic phenotype of CaSki cells, as Notch silencing was growth inhibitory [19]. On the other hand, exaggerated levels of Notch had detrimental effects on growth and survival of HeLa [6, 18], similar to our findings in SiHa cells. How Notch expression and activity is regulated is not completely understood. Several lines of evidence indicate that HPV E6E7 expression may upregulate Notch expression in primary cells [6, 19] as was also seen in our previous study showing increased Notch mRNA expression in HPV-immortalized cells compared to primary keratinocytes [20]. On the other hand E6E7 expression itself also appears to be regulated by Notch, most likely mediated by AP-1 [6, 16, 18]. Additionally, Notch activity is known to depend on ligand availability on neighboring cells (reviewed by [1, 8]). Interestingly, both Jagged and Delta-like 1 ligands, which regulate Notch signaling, show the same expression profile as Notch at the different stages of transformation, i.e. increased in HPV-immortalized cells and reduced in cancer cell lines (data not shown), and may therefore contribute to Notch regulation in the HPV-containing cells.
Based on present findings and literature data we conclude that during early stages of transformation, i.e. from mortal primary cells, to HPV-immortalized cells and anchorage independent cells, Notch expression rises, to aid the early steps in transformation, while after accomplishment of transformation Notch expression is downregulated, apparently to stimulate events inducing tumorigenicity. We furthermore demonstrated that Notch functioning is dose dependent, with a moderate dose inducing transformation and a high dose inducing a change in AP-1 composition in favour of cJun/Fra1 dimers, leading to reduction of anchorage independent growth.
References
E.J. Allenspach, I. Maillard, J.C. Aster, W.S. Pear, Notch signaling in cancer. Cancer Biol. Ther. 1, 466–476 (2002)
A. Bachmann, B. Hanke, R. Zawatzky, U. Soto, J. van Riggelen, H. zur Hausen, F. Rosl, Disturbance of tumor necrosis factor alpha-mediated beta interferon signaling in cervical carcinoma cells. J. Virol. 76, 280–291 (2002)
B. Daniel, A. Rangarajan, G. Mukherjee, E. Vallikad, S. Krishna, The link between integration and expression of human papillomavirus type 16 genomes and cellular changes in the evolution of cervical intraepithelial neoplastic lesions. J. Gen. Virol. 78, 1095–1101 (1997)
J. De-Castro Arce, U. Soto, J. van Riggelen, E. Schwarz, H. zur Hausen, F. Rosl, Ectopic expression of nonliganded retinoic acid receptor beta abrogates AP-1 activity by selective degradation of c-Jun in cervical carcinoma cells. J. Biol. Chem. 279, 45408–45416 (2004)
R. Eferl, E.F. Wagner, AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer. 3, 859–868 (2003)
S. Lathion, J. Schaper, P. Beard, K. Raj, Notch1 can contribute to viral-induced transformation of primary human keratinocytes. Cancer Res. 63, 8687–8694 (2003)
T.T. Maliekal, J. Bajaj, V. Giri, D. Subramanyam, S. Krishna, The role of Notch signaling in human cervical cancer: implications for solid tumors. Oncogene 27, 5110–5114 (2008)
F. Radtke, K. Raj, The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer. 3, 756–767 (2003)
B. Ramdass, T.T. Maliekal, S. Lakshmi, M. Rehman, P. Rema, P. Nair, G. Mukherjee, B.K. Reddy, S. Krishna, P.M. Radhakrishna, Coexpression of Notch1 and NF-kappaB signaling pathway components in human cervical cancer progression. Gynecol. Oncol. 104, 352–361 (2007)
A. Rangarajan, R. Syal, S. Selvarajah, O. Chakrabarti, A. Sarin, S. Krishna, Activated Notch1 signaling cooperates with papillomavirus oncogenes in transformation and generates resistance to apoptosis on matrix withdrawal through PKB/Akt. Virology 20(286), 23–30 (2001)
U. Soto, B.C. Das, M. Lengert, P. Finzer, H. zur Hausen, F. Rosl, Conversion of HPV 18 positive non-tumorigenic HeLa-fibroblast hybrids to invasive growth involves loss of TNF-alpha mediated repression of viral transcription and modification of the AP-1 transcription complex. Oncogene 18, 3187–3198 (1999)
U. Soto, C. Denk, P. Finzer, K.J. Hutter, H. zur Hausen, F. Rosl, Genetic complementation to non-tumorigenicity in cervical-carcinoma cells correlates with alterations in AP-1 composition. Int. J. Cancer 86, 811–817 (2000)
R.D. Steenbergen, D. Kramer, B.J. Braakhuis, P.L. Stern, R.H. Verheijen, C.J. Meijer, P.J. Snijders, TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl. Cancer Inst. 96, 294–305 (2004)
R.D. Steenbergen, J.M. Walboomers, C.J. Meijer, E.M. van der Raaij-Helmer, J.N. Parker, L.T. Chow, T.R. Broker, P.J. Snijders, Transition of human papillomavirus type 16 and 18 transfected human foreskin keratinocytes towards immortality: activation of telomerase and allele losses at 3p, 10p, 11q and/or 18q. Oncogene 19;13, 1249–1257 (1996)
C. Talora, S. Cialfi, O. Segatto, S. Morrone, C.J. Kim, L. Frati, D.G. Paolo, A. Gulino, I. Screpanti, Constitutively active Notch1 induces growth arrest of HPV-positive cervical cancer cells via separate signaling pathways. Exp. Cell Res. 305, 343–354 (2005)
C. Talora, D.C. Sgroi, C.P. Crum, G.P. Dotto, Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Dev. 16, 2252–2263 (2002)
F. Thierry, G. Spyrou, M. Yaniv, P. Howley, Two AP1 sites binding JunB are essential for human papillomavirus type 18 transcription in keratinocytes. J. Virol. 66, 3740–3748 (1992)
L. Wang, H. Qin, B. Chen, X. Xin, J. Li, H. Han, Overexpressed active Notch1 induces cell growth arrest of HeLa cervical carcinoma cells. Int J Gynecol. Cancer. 17, 1283–1292 (2007)
S. Weijzen, A. Zlobin, M. Braid, L. Miele, W.M. Kast, HPV16 E6 and E7 oncoproteins regulate Notch-1 expression and cooperate to induce transformation. J. Cell. Physiol. 194, 356–362 (2003)
J. de Wilde, J. De-Castro Arce, P.J. Snijders, C.J. Meijer, F. Rosl, R.D. Steenbergen, Alterations in AP-1 and AP-1 regulatory genes during HPV-induced carcinogenesis. Cell. Oncol. 30, 77–87 (2008)
P. Zagouras, S. Stifani, C.M. Blaumueller, M.L. Carcangiu, S. Artavanis-Tsakonas, Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc. Natl. Acad. Sci. U.S.A. 92, 6414–6418 (1995)
Acknowledgements
The authors are thankful for excellent technical assistance to Chantal Stoepker, Jeroen Paardekooper-Overman and Elke Gockel.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Henken, F.E., De-Castro Arce, J., Rösl, F. et al. The functional role of Notch signaling in HPV-mediated transformation is dose-dependent and linked to AP-1 alterations. Cell Oncol. 35, 77–84 (2012). https://doi.org/10.1007/s13402-011-0062-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-011-0062-4