Abstract
Background
Expression of estrogen receptor alpha (ERα) is predictive for endocrine therapy response and an important prognostic factor in breast cancer. Overexpression of ERα can be caused by estrogen receptor 1 (ESR1) gene amplification and was originally reported to be a frequent event associated with a significantly longer survival for ER-positive women treated with adjuvant tamoxifen monotherapy, which was however questioned by subsequent studies.
Methods
This study aimed to reanalyze the frequency of ESR1 amplification by multiplex ligation-dependent probe amplification (MLPA) and fluorescence in situ hybridisation (FISH), and to assess clinicopathologic correlations. MLPA was performed in a group of 135 breast cancer patients, and gains/amplifications were subjected to FISH.
Results
True ESR1 amplification by MLPA was rare (2%) and only 6% more patients showed a modest gain of ESR1. All MLPA-detected ESR1 amplifications and nearly all ESR1 gains were also FISH amplified and gained, but not all FISH amplifications/gains were MLPA amplified/gained, leading to an overall concordance of only 60% between both techniques. All 3 MLPA and FISH ESR1 amplified cases had high ERα expression, but there was no obvious correlation between ESR1 gain and ER status by IHC. ESR1 gains/amplifications were not associated with HER2 gain/amplification, but seemed to be associated with older age. Surprisingly, ESR1 gain/amplification was not associated with low grade as reported previously, but correlated with high grade and high proliferation. Furthermore, ESR1 gain/amplification by MLPA was not associated with nodal status or tumor size (pT status).
Conclusions
ESR1 amplification as detected by MLPA is rare in breast cancer, and seems to be associated with high ERα expression, high age, high grade and high proliferation. This study confirms previous studies that showed differences in the ESR1 amplification frequencies detected by different techniques.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The choice of therapy for breast cancer is based on clinico-pathological features such as estrogen receptor (ERα) and progesterone receptor (PR) status, HER2 (human epidermal growth factor receptor 2) status, tumor size and grade. Expression of ERα is predictive for endocrine therapy response and an important prognostic factor for breast cancer [1]. ERα is a member of the nuclear hormone-receptor superfamily. The gene coding for this receptor, ESR1 (estrogen receptor 1), is located on chromosome 6q25 [2]. ERα regulates the transcription of many genes (approximately 5% of the genome) by binding to estrogen responsive elements (ERE) thereby leading to e.g. cell proliferation. Estrogen and ERα are involved in sexual development and reproductive function, but in pathological situations elevated levels of ERα are seen. Ductal carcinoma in situ of the breast generally expressess ERα, and more than two-thirds of invasive breast cancers are ERα positive [3]. Several endocrine therapies targeting ERα have successfully been developed, leading to a substantial decrease in tumor growth in about 30–50% of ERα expressing patients [4]. Tamoxifen is a selective estrogen receptor modulator (SERM) and is the standard endocrine therapy for ER-positive breast cancers [4]. It competes with estrogen for binding to ERα and thereby inhibits gene transcription activation and thus cell growth. Other endocrine therapies that are increasingly used in breast cancer treatment are aromatase inhibitors (AIs), which block the synthesis of estrogen [5] and fulvestrant, an estrogen-receptor destabilizator and downregulator (SERD) [6].
Overexpression of ERα can be caused by ESR1 amplification and ESR1 amplification by fluorescence in situ hybridization (FISH) was originally reported to be a relatively frequent event in breast cancer, associated with a significantly longer survival for patients treated with adjuvant tamoxifen monotherapy [7]. Amplification of ESR1 has subsequently been studied by several other groups that have questioned the frequency of ERα amplification in breast cancer patients [8]. ERα gene amplification detection using FISH provided a frequency of 20.6% [7] and 22.6% [9], whereas aCGH, CISH and qPCR provided a frequency of 0–10% [10–13]. Differences in amplification frequencies were assigned to missing the amplification due to the small amplicon (600 kb) and the lower levels of amplification, to tumor heterogeneity and to contamination by normal cells. The latter two would lead to an underestimation of the amplification frequency detected by non-morphological techniques. Other confounding factors might be the different interpretation of the definition ‘amplification’ and/or the use of an automated/manual scoring system for ISH [8].
Multiplex ligation-dependent probe amplification (MLPA) is a molecular technique to detect gene copy number changes, that, for HER2, has been shown to correlate well with immunohistochemistry (IHC), fluorescence in situ hybridization (FISH) and chromogenic in situ hybridization (CISH) [14]. MLPA is a PCR-based method for gene copy number quantification in DNA extracted from frozen or paraffin embedded tissue. MLPA kits contain up to 45 probes which can be simultaneously detected in one PCR reaction [15, 16]. This study aimed to analyze the frequency of ESR1 amplification by MLPA in a large group of breast cancer patients and to compare ESR1 gains and amplifications detected by MLPA with FISH and clinicopathologic features. This study will show that ESR1 amplification detected by MLPA is rare in breast cancer, and seems to be associated with high ERα expression, high age, high grade and high proliferation.
2 Materials and methods
2.1 Patient material
Tissue samples of invasive breast cancer patients were collected between November 2004 and September 2009 at the Department of Pathology of the University Medical Centre in Utrecht (UMCU), The Netherlands. This study randomly selected 135 tissue samples from this consecutive series. According to the Dutch Federation of Medical Scientific Societies (the FEDERA), use of redundant tissue for research purposes does not require informed consent, if the patients are offered the possibilities to refuse this (“opting out” system) and material has been used anonymously or coded. In the UMC Utrecht, all patients are informed that research may happen with their redundant tissue, and are offered to opt out [17]. No material of patients that have opted out has been used in the present study, and all materials were used anonymously. The research protocol for this study was approved by the Scientific Advisory Counsel of the UMC Utrecht Biobank. Grading was performed according to the Nottingham modification of Bloom-Richardson system, and mitoses were counted according to a strict protocol as before to arrive at the mitotic activity index (MAI) [18]. All tissue samples were analyzed with MLPA to determine ESR1 and HER2 gene copy number alternations. Furthermore, HER2, ER and PR protein expression were assessed by immunohistochemistry, and histological type, tumor size, histological grade and age at diagnosis were determined for all patients. Tumors showing ESR1 gain or amplification by MLPA were re-analyzed by FISH, as were 14 tumors with normal MLPA ESR1 results. For all FISH-analyzed tumors in this study, HER2 CISH data were already available from previous studies [14, 19].
2.2 Immunohistochemistry
IHC for HER2 was performed using the Hercep test (Dako, Glostrup, Denmark) according to the manufacturers’ instructions on 4 μm thick sections from the neutral buffered formaldehyde fixed tissue blocks. IHC membrane staining was semi-quantitatively scored as negative (0), weakly positive (1+), equivocal (2+) and strongly positive (3+) according to the DAKO FDA-approved scoring system. Interpretation of staining was done by 2 experienced breast pathologists. As control a small tissue array containing a 0, 1+, 2+ and 3+ breast tumor samples was taken along on the same slide as the tumor to be analyzed. Immunohistochemical staining for ER (1D5, 1:80, Dako) and PR (PGR636, 1:200, Dako) was performed using a Bond-Max automated staining machine (Vision Biosystems, Newcastle, UK) with the Bond polymer refine detection kit (Vision BioSystems, cat. no DS9800). Appropriate positive and negative controls were used throughout.
2.3 Multiplex ligation-dependent probe amplification (MLPA)
Invasive tumor areas as identified on serial H&E sections were harvested from one or two whole 4 μm thick paraffin sections (corresponding to approximately 1 square cm of tumor tissue) with a scalpel. DNA was isolated from these tissue fragments by 1 hr incubation in proteinase K (10 mg/ml; Roche, Almere, The Netherlands) and lysis buffer (50 mM Tris-HCl buffer pH 8.0 with 0.5% Tween 20) at 56°C followed by boiling for 10 min. This DNA solution (50–100 μl) was, after centrifugation, used in the MLPA analysis according the manufacturers’ instructions, using the P004-B1 kit (MRC Holland, Amsterdam, The Netherlands). This kit contains, amongst others, two probes for ESR1 and three for HER2. All tests were performed in duplicate in an ABI 9700 PCR machine (Applied Biosystems, Foster City, CA, USA). PCR products were analyzed on an ABI310 capillary sequencer (Applied Biosystems). Gene copy numbers were analyzed using Genescan (Applied Biosystems) and Coffalyser (version 9.4) software (MRC-Holland). For genes with more than one probe present in the kit, the mean of all the probe peaks of this gene in duplicate was calculated. If this mean value was below 0.7 the respective gene was defined as lost, a value between 0.7–1.3 was defined as normal, 1.3–2.0 as gain, and values >2.0 as amplified, as previously established [20, 21].
2.4 FISH
FISH was performed using the ZytoLight®SPEC ESR1/CEN 6 Dual Color Probe kit (Zytovision, Germany, Z-2070-20) according to the manufacturer’s instructions. Briefly, slides were deparaffinised and incubated for 15 min in Heat Pretreatment Solution Citric at 98°C. Slides were incubated in a pepsin solution for 10 min at 37º C, washed in Wash buffer SSC, dehydrated and air dried. Subsequently, 10 μl of ZytoLight®SPEC ESR1/CEN 6 Dual Color Probe was applied to the slides followed by denaturation at 75°C for 10 min and incubation overnight at 37°C in a Thermobrite StatSpin system (Abbott Molecular). After hybridization, coverslips were removed in Wash Buffer A at 37°C for 2 min, followed by a wash in the same Wash Buffer for 2× 5 min at 37°C, dehydration and DAPI/antifade solution for 15 min in the dark. Evaluation of the sample was carried out using a Leica DM5500 B fluorescence microscope (Leica Microsystems, Germany) and Leica AF6000 software. The ESR1 probe was labelled with ZyGreen (green, excitation at 503 nm and emission at 528 nm) and the CEP6 alpha-satellite probe with ZyOrange (orange, excitation at 547 nm and emission at 572 nm). Interpretation of the results was based on the counting of at least 30 tumor cells in at least two different areas by one blinded observer, and we used two different approaches: the ESR1 absolute copy number and the ESR1/CEP6 ratio. A sample was defined as gained if the absolute ESR1 copy number was >2 or the ESR1/CEP6 ratio was >1.0, and as amplified if the absolute ESR1 copy number was >10 or if the ESR1/CEP6 ratio was >2.2.
3 Results
3.1 MLPA
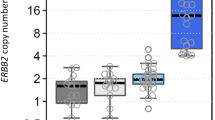
Of the 135 tumors analyzed by MLPA, 2% showed ESR1 amplification (3/135) and 6% showed ESR1 gain (8/135). Fifteen percent of the patients showed HER2 amplification (20/135) by MLPA, and 5% more showed HER2 gain (7/135). None of the 3 true ESR1 amplifications were associated with HER2 amplification, but 4/8 ESR1 gains were associated with HER2 gain (2/4) or HER2 amplification (2/4). There were no losses of ESR1 by MLPA in this study, although 10 tumors had copy number ratios in the lower range between 0.7 and 0.8.
3.2 FISH
Supplementary Table 1 shows the association between ESR1 copy number status by MLPA and ESR1 status by FISH (separated into the ESR1/CEP6 ratio and the ESR1 absolute copy number). Figure 1 and Tables 1 and 2 show that all three amplifications by MLPA were also amplified by FISH using both FISH interpretation methods. Table 1 shows that four of the 8 MLPA-detected ESR1 gains were also FISH amplified, 2/8 were FISH gained and 2/8 were not amplified by FISH using the ratio interpretation. Of all 25 FISH-analyzed samples, 13/25 (52%) showed an increased (>2) CEP6 copy number.
Using the absolute copy number interpretation as shown in Table 2, 3/8 MLPA-detected gains were also FISH amplified, 4/8 were FISH gained and 1/8 MLPA gains was not amplified by FISH.
We also analyzed 14 MLPA non-amplified breast tumors by FISH of which 10/14 were normal, 1/14 was gained and 3/14 were amplified by FISH using the ratio interpretation. Using the absolute copy number interpretation, 6/14 MLPA non-amplified tumors showed no amplification by FISH, 7/14 were gained and 1/14 was amplified by FISH. This study found two ESR1 losses by FISH which were both associated with lower MLPA copy number ratios (0.72 and 0.73).
3.3 Association with clinicopathological features
Supplementary Table 1 shows that all three MLPA-detected ESR1 amplifications were associated with 100% ER positivity. Of the eight ESR1 gains by MLPA, 5 had very high ER expression (90–100%) and 3 were ER negative. Two of the 3 tumors with ESR1 amplifications were grade 3 and 1 was grade 1. All 8 tumors with ESR1 gains were non-low grade (5/8 grade 3, 3/8 grade 2) and, in total, 24/25 grade 1 tumors had normal ESR1 gene dosage. Of the three ESR1 amplifications and eight ESR1 gains, 2/3 and 6/8 had high MAI (>10), respectively. All but one (lobular) tumors with ESR1 amplifications or gains were of the ductal type. All patients with ESR1 gain/amplification by MLPA were older than 50 years. Two of the three MLPA-detected amplifications were pT1, the other was pT2. Six of the 8 ESR1 gains were pT1 (<2 cm), whereas 2/8 were pT2. The lymph node status of the 3 patients with MLPA-detected amplifications was pN0 in 2/3, and pN1 in 1/3. For the MLPA-detected gains, 5/8 were pN0 and 3/8 pN1.
Table 1 shows 10 ESR1 FISH amplifications and 3 gains using the ESR1/CEP6 ratio for interpretation. All 10 FISH amplifications had high ER expression (90–100%) but 2/3 gains were completely ER negative. None of the ESR1 FISH amplified tumors were HER2 CISH amplified, but all 3 FISH-detected gains were HER2 CISH amplified. Nine of the 10 FISH amplifications were non-low grade (7/9 grade 3, 2/9 grade 2). All but two of these ESR1 FISH amplifications were also associated with high MAI. Five and 6 of the 10 ESR1 FISH amplifications were pN0 and pT1 respectively. The other five and 4 ESR1 amplifications were pN1 and pT2, respectively. Two patients with ESR1 FISH amplification were younger than 50. All three gains were non-low grade (1/3 grade 3, 2/3 grade 2), had a high MAI, were pN0, pT1 and older than 50.
Using the absolute ESR1 copy number for interpretation, all 7 FISH amplifications had high ER expression (90–100%) but only 5/11 FISH gains were ER positive. None of the ESR1 FISH amplified tumors were HER2 CISH amplified, but 3/11 FISH-detected gains were HER2 CISH amplified. Five of the seven amplifications were non-low grade (4/5 grade 3 and 1/5 grade 2), and all but two of the amplifications were associated with high MAI. Of the 7 amplifications, 4/7 were pN0 and 3/7 pN1, 3/7 were pT1 and 4/7 pT2. All 7 patients with ESR1 amplifications were older than 50 years of age. All but one of the 11 FISH-detected gains were non-low grade (7/10 grade 3 and 3/10 grade 2) and 9/11 were associated with high MAI. Of the 11 detected ESR1 gains, 5/11 were pN0, 4 were pN1 and 2 were pN2. Seven of these 11 gains were associated with pT1, 3/11 with pT2 and 1/11 was pT3. Five of the 11 patients with FISH-detected ESR1 gains were younger than 50.
4 Discussion
The aim of this study was to analyze the frequency of ESR1 amplification by MLPA, to confirm these amplifications by FISH, and to associate these amplifications with clinico-pathological features. ESR1 amplification by MLPA was rare (2%) and additionally 6% of the patients showed ESR1 gain. All MLPA-detected ESR1 amplifications and nearly all ESR1 gains were also FISH amplified and gained, but not all FISH amplifications/gains were MLPA amplified/gained, with a 60% overall concordance (15/25) between MLPA and FISH using the ratio interpretation method and a 52% concordance (13/25) between MLPA and FISH using the absolute ESR1 copy number. Although the ESR1/CEP17 FISH ratio showed a slightly better correlation with MLPA than the absolute FISH ESR1 copy number, there was overall not a very good concordance between MLPA and FISH. FISH seemed to detect more amplifications than MLPA (in this selected group 40% (10/25) plus 12% (3/25) gains). Our percentage of ESR1 amplification by MLPA was much lower than the 20.6% found by Holst et al. [7] using BAC FISH on tissue microarrays, but was consistent with the lower range (0–10%) of ESR1 amplification reported by other groups using a broad range of techniques such as array comparative genomic hybridization (aCGH), CISH, FISH and qPCR [8–13, 22]. Several reasons have been postulated for the difference in amplification frequencies reported by these techniques including heterogeneity or contamination by normal DNA (aCGH, qPCR, MLPA in this study—although we microdissected the tumor area), the small ESR1 amplicon size (although HER2 also has a small amplicon size: 500–700 kb), low levels of amplification (only 15% of its amplifications were >10 copies, 41% 5–6 copies), large CISH probes (360 kb), and an automated/manual scoring system. According to Holst et al. [22], detection of ESR1 amplification by FISH will require modifications of the established procedures.
Given the low amount of ESR1 gains and amplifications in this study, it was not deemed appropriate to perform statistical analysis. Therefore, for the association with clinicopathological parameters, frequencies were used rather than statistical p-values. ESR1 gain/amplification was not associated with low grade as reported by Holst et al. [7], but in contrary, seemed to be associated with higher grade (2 or 3) and high MAI. This could however explain in part the observation of Ejlertsen et al. that patients with ESR1 amplification treated with tamoxifen had a shorter time to recurrence [23] and the fact that Lyng et al. [24] found ESR1 amplification more frequently in patients with recurrence (6/7 amplified patients showed recurrence in contrast to 22/46 patients without ESR1 amplification) upon tamoxifen treatment. We did not find an obvious relation between ESR1 gain/amplification and tumor size (pT status) or lymph node status (pN status), but ESR1 gains/amplifications seemed to be associated with higher age.
Similar to Holst et al., all ESR1 amplifications detected by MLPA or FISH were 90–100% ERα positive. For ESR1 gains, there was no obvious correlation with ERα protein overexpression.
5 Conclusion
In conclusion, this study used MLPA to detect ESR1 amplification in a group of 135 patients and detected only 2% amplification and 6% more gains. All ESR1 amplifications by MLPA or FISH had high ERα expression by IHC, but for ESR1 gains this was not obvious. ESR1 gains/amplifications were not associated with HER2 gain/amplification. Surprisingly, ESR1 gain/amplification was not preferably associated with low-grade tumors as reported previously, but in contrast seemed to be associated with high grade and high MAI. ESR1 amplification by MLPA was associated with higher age, but not with nodal status or pT status. All MLPA-detected ESR1 amplifications and nearly all ESR1 gains were also FISH amplified and gained, but not all FISH amplifications/gains were MLPA amplified/gained, leading to an overall concordance of only 60% between both techniques. This confirms previous studies that showed differences in the amplification frequencies detected by different techniques.
References
S.I. Hayashi, H. Eguchi, K. Tanimoto, T. Yoshida, Y. Omoto, A. Inoue et al., The expression and function of estrogen receptor alpha and beta in human breast cancer and its clinical application. Endocr Relat Cancer 10, 193–202 (2003)
L.P. Menasce, G.R. White, C.J. Harrison, J.M. Boyle, Localization of the estrogen receptor locus (ESR) to chromosome 6q25.1 by FISH and a simple post-FISH banding technique. Genomics 17, 263–265 (1993)
D.C. Allred, S.K. Mohsin, Biological features of premalignant disease in the human breast. J. Mammary Gland Biol. Neoplasia 5, 351–364 (2000)
C.X. Ma, C.G. Sanchez, M.J. Ellis, Predicting endocrine therapy responsiveness in breast cancer. Oncology (Williston Park) 23, 133–142 (2009)
A. Howell, A. Buzdar, Are aromatase inhibitors superior to antiestrogens? J. Steroid Biochem. Mol. Biol. 93, 237–247 (2005)
A.E. Wakeling, Similarities and distinctions in the mode of action of different classes of antioestrogens. Endocr. Relat. Cancer 7, 17–28 (2000)
F. Holst, P.R. Stahl, C. Ruiz, O. Hellwinkel, Z. Jehan, M. Wendland et al., Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat. Genet. 39, 655–660 (2007)
D.G. Albertson, Conflicting evidence on the frequency of ESR1 amplification in breast cancer. Nat. Genet. 40, 821–822 (2008)
S. Tomita, Z. Zhang, M. Nakano, M. Ibusuki, T. Kawazoe, Y. Yamamoto et al., Estrogen receptor alpha gene ESR1 amplification may predict endocrine therapy responsiveness in breast cancer patients. Cancer Sci. 100, 1012–1017 (2009)
L.A. Brown, J. Hoog, S.F. Chin, Y. Tao, A.A. Zayed, K. Chin et al., ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. 40, 806–807 (2008)
J.S. Reis-Filho, S. Drury, M.B. Lambros, C. Marchio, N. Johnson, R. Natrajan et al., ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. 40, 809–810 (2008)
H.M. Horlings, A. Bergamaschi, S.H. Nordgard, Y.H. Kim, W. Han, D.Y. Noh et al., ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. 40, 807–808 (2008)
A. Vincent-Salomon, V. Raynal, C. Lucchesi, N. Gruel, O. Delattre, ESR1 gene amplification in breast cancer: a common phenomenon? Nat. Genet. 40, 809–2 (2008)
C.B. Moelans, R.A.de Weger, M.T.van Blokland, C. Ezendam, S. Elshof, M.G. Tilanus et al., HER-2/neu amplification testing in breast cancer by multiplex ligation-dependent probe amplification in comparison with immunohistochemistry and in situ hybridization. Cell. Oncol. 31, 1–10 (2009)
C.B. Moelans, R.A.de Weger, M.T.van Blokland, W.E.van der, and P.J.van Diest, Simultaneous detection of TOP2A and HER2 gene amplification by multiplex ligation-dependent probe amplification in breast cancer. Mod. Pathol. 23, 62–70 (2010)
C.B. Moelans, R.A.de Weger, and P.J.van Diest, Absence of chromosome 17 polysomy in breast cancer: analysis by CEP17 chromogenic in situ hybridization and multiplex ligation-dependent probe amplification. Breast Cancer Res. Treat. 120, 1–7 (2010)
P.J. van Diest, No consent should be needed for using leftover body material for scientific purposes. BMJ 325, 648–651 (2002)
P.J. van Diest, J.P. Baak, P. Matze-Cok, E.C. Wisse-Brekelmans, C.M. van Galen, P.H. Kurver et al., Reproducibility of mitosis counting in 2,469 breast cancer specimens: results from the Multicenter Morphometric Mammary Carcinoma Project. Hum. Pathol. 23, 603–607 (1992)
C.B. Moelans, R.A. de Weger, C. Ezendam, P.J. van Diest, HER-2/neu amplification testing in breast cancer by Multiplex Ligation-dependent Probe Amplification: influence of manual—and laser microdissection. BMC Cancer 9, 4 (2009)
D.J. Bunyan, D.M. Eccles, J. Sillibourne, E. Wilkins, N.S. Thomas, J. Shea-Simonds et al., Dosage analysis of cancer predisposition genes by multiplex ligation-dependent probe amplification. Br. J. Cancer 91, 1155–1159 (2004)
J. Coffa, M.A.van de Wiel, B. Diosdado, B. Carvalho, J. Schouten, and G.A. Meijer, MLPAnalyzer: data analysis tool for reliable automated normalization of MLPA fragment data. Cell. Oncol. 30, 323–335 (2008)
F. Holst, P. Stahl, O. Hellwinkel, A.-M. Dancau, A. Krohn, L. Wuth et al., Reply to: ESR1 gene amplification in breast cancer: a common phenomenon? Nature 40, 810–812 (2008)
B. Ejlertsen, K.V. Nielsen, B.B. Rasmussen, E. Balslev, S. Muller, H.T. Mouridsen, Amplification of ESR1 may predict resistance to adjuvant tamoxifen in postmenopausal patients with hormone receptor positive breast cancer. Breast Cancer Res. Treat. 106, S34-abstract 402 (2007)
M.B. Lyng, K. Vang, A. Knoop, H. Ditzel, A.E. Lykkesfeldt, S. Muller et al., ER Status Measured by FISH Is a Better Predictor of Prognosis in ER Positive, Postmenopausal Breast Cancer Patients Treated with Adjuvant Tamoxifen Than ER Status Measured by RT-qPCR or IHC, San Antonio Breast Cancer Symposium (2009), Poster # 2004
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper is a reprint from ‘ESR1 Amplification is rare in Breast Cancer and is associated with high Grade and high Proliferation: a multiplex ligation-dependent probe amplification Study, Cathy B Moelans, Hanneke N Monsuur, Johannes H de Pinth, Remco D Radersma, Roel A de Weger and Paul J van Diest’ originally published in Analytical Cellular Pathology/Cellular Oncology, Volume 33, number1, 2010, pp. 13–18, IOS Press.
An erratum to this article can be found at http://dx.doi.org/10.1007/s13402-011-0063-3
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
ESR1 status by multiplex ligation-dependent probe amplification (MLPA) and fluorescence in situ hybridisation (FISH, according to the ratio ESR1/CEP6 and the absolute ESR1 copy number) for 25 breast cancer patients. (DOC 100 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Moelans, C.B., Monsuur, H.N., de Pinth, J.H. et al. ESR1 amplification is rare in breast cancer and is associated with high grade and high proliferation: a multiplex ligation-dependent probe amplification study. Cell Oncol. 34, 489–494 (2011). https://doi.org/10.1007/s13402-011-0045-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-011-0045-5