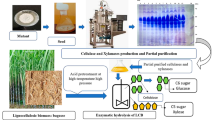
Abstract
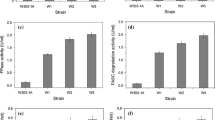
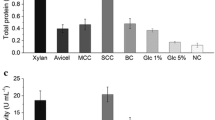
An indigenous thermophilic fungal strain of Mycothermus thermophilus (Syn. Scytalidium thermophilum) designated as CM 4 T was subjected to a combination of cyclic mutagenesis and intra-specific protoplast fusion to induce heterokaryosis and diploidization for developing deregulated cellulase hyper-producer strain. The developed strain (23 U) produced 14.60, 45.50, 82.80, 3.28, and 264 U/ml of CBH, β-glucosidase, CMCase, Fpase, and xylanase at fermenter level, which were 8.6, 2.54, 6.58, 2.37, and 6.58 folds improved, respectively, in comparison to wild-type strain CM 4 T. Furthermore, the developed mutants produced cellulases constitutively during fed-batch (flask mode) using glucose feed at 0.375 g carbon/l/h. The creA gene and CreA protein structures of mutant 23 U showed truncated zinc finger motif linking it to the catabolite repression-resistant phenotype. The hydrolytic potential of cellulase produced of developed strain 23 U was evaluated using acid- and alkali-treated rice straw and bagasse at 10 % substrate loading rate. Furthermore, studies showed that hydrolytic efficiency of the cellulases produced by mutant 23 U can be further enhanced by custom designing of the cocktails that comprised of lignocellulolytic enzymes of mutant 23 U and different thermophilic/thermotolerant fungal strains.






Similar content being viewed by others
References
Liu G, Zhang J, Bao J (2016) Cost evaluation of cellulase enzyme for industrial-scale cellulosic ethanol production based on rigorous Aspen Plus modeling. Bioprocess Biosyst Eng 39:133–140. https://doi.org/10.1007/s00449-015-1497-1
Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VGH (2012) Novel enzymes for the degradation of cellulose. Biotechnol Biofuels 5:45. https://doi.org/10.1186/1754-6834-5-45
Knott BC, Crowley MF, Himmel ME, Ståhlberg J, Beckham GT (2014) Carbohydrate-protein interactions that drive processive polysaccharide translocation in enzymes revealed from a computational study of cellobiohydrolase processivity. J Am Chem Soc 136:8810–8819
Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, Sandgren M, Ståhlberg J, Beckham GT (2015) Fungal cellulases. Chem Rev 115:1308–1448. https://doi.org/10.1021/cr500351c
Rarbach M, Dragovic Z, Kohl A, Gerlach J, Bartuch J, Brück T (2014) Efficient lignocellulose hydrolysis with integrated enzyme production. U.S. Patent US 2014/0045227 A1.
Johansen KS (2016) Discovery and industrial applications of lytic polysaccharide mono-oxygenases. Biochem Soc Trans 44:143–149. https://doi.org/10.1042/BST20150204
Visser H, Joosten V, Punt PJ, Gusakov AV, Olson PT, Joosten R et al (2011) Development of a mature fungal technology and production platform for industrial enzymes based on a Myceliophthora thermophila isolate, previously known as Chrysosporium lucknowense C1. Ind Biotechnol 7:214–223. https://doi.org/10.1089/ind.2011.7.214
Bevers L E, Appeldoorn M, Schooneveld-Bergmans M E F, Pel H J (2019) Enzyme Composition. US Patent Apllication 2019/0276809 A1.
Gabriel R, Prinz J, Jecmenica M, Romero-Vazquez C, Chou P, Harth S, Floerl L, Curran L, Oostlander A, Matz L, Fritsche S, Gorman J, Schuerg T, Fleißner A, Singer SW (2020) Development of genetic tools for the thermophilic filamentous fungus Thermoascus aurantiacus. Biotechnol Biofuels 13:167. https://doi.org/10.1186/s13068-020-01804-x
Mahajan C, Chadha BS, Nain L, Kaur A (2014) Evaluation of glycosyl hydrolases from thermophilic fungi for their potential in bioconversion of alkali and biologically treated Parthenium hysterophorus weed and rice straw into ethanol. Bioresour Technol 163:300–307. https://doi.org/10.1016/j.biortech.2014.04.057
Basotra N, Kaur B, Di Falco M et al (2016) Mycothermus thermophiles (Syn. Scytalidium thermophilum): Repertoire of a diverse array of efficient cellulases and hemicellulases in the secretome revealed. Bioresour Technol 222:413–421. https://doi.org/10.1016/j.biortech.2016.10.018
Kaur B, Oberoi HS, Chadha BS (2016) Enhanced cellulase producing mutants developed from heterokaryotic Aspergillus strain. Bioresour Technol 156:100–107. https://doi.org/10.1016/j.biortech.2014.01.016
Kaur B, Sharma M, Soni R, Oberoi HS, Chadha BS (2013) Proteome-based profiling of hypercellulase-producing strains developed through interspecific protoplast fusion between aspergillus nidulans and aspergillus tubingensis. Appl Biochem Biotechnol 169:393–407. https://doi.org/10.1007/s12010-012-9985-0
Kaur R, Chadha BS, Singh S, Saini HS (2000) Amylase hyper-producing haploid recombinant strains of Thermomyces lanuginosus obtained by intra-specific protoplast fusion. Can J Microbiol 46:669–673. https://doi.org/10.1139/cjm-46-7-669
Han X, Song W, Liu G et al (2017) Improving cellulase productivity of Penicillium oxalicum RE-10 by repeated fed batch fermentation strategy. Bioresor Technol. https://doi.org/10.1016/j.biortech.2016.11.079
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. https://doi.org/10.1038/nprot.2015.053
Capra JA, Singh M (2007) Predicting functionally important residues from sequence conservation. Bioinformatics 23:1875–1882. https://doi.org/10.1093/bioinformatics/btm270
Brar KK, Agrawal D, Chadha BS, Lee H (2019) Evaluating novel fungal secretomes for efficient saccharification and fermentation of composite sugars derived from hydrolysate and molasses into ethanol. Bioresour Technol 273:114–121. https://doi.org/10.1016/j.biortech.2018.11.004
Badhan AK, Chadha BS, Kaur J, Saini HS, Bhat MK (2007) Production of multiple xylanolytic and cellulolytic enzymes by thermophilic fungus Myceliophthora sp. IMI 387099. Bioresour Technol 98:504–510. https://doi.org/10.1016/j.biortech.2006.02.009
Brar KK, Kaur S, Chadha BS (2016) A novel staggered hybrid SSF approach for efficient conversion of cellulose/hemicellulosic fractions of corncob into ethanol. Renew Energy 98:16–22. https://doi.org/10.1016/j.renene.2016.03.082
Breslmayr E, Hanžek M, Hanrahan A, Leitner C, Kittl R, Šantek B, Oostenbrink C, Ludwig R (2018) A fast and sensitive activity assay for lytic polysaccharide monooxygenase. Biotechnol Biofuels 11:79. https://doi.org/10.1186/s13068-018-1063-6
Miller GL (1959) Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal Chem 31:426–428. https://doi.org/10.1021/ac60147a030
Basotra N, Joshi S, Satyanarayana T, Pati PK, Tsang A, Chadha BS (2018) Expression of catalytically efficient xylanases from thermophilic fungus Malbranchea cinnamomea for synergistically enhancing hydrolysis of lignocellulosics. Int J Biol Macromol 108:185–192. https://doi.org/10.1016/j.ijbiomac.2017.11.131
Adney B, Baker J (2008) Measurement of Cellulase Activities. Laboratory Analytical Procedure (LAP), Issue Date, 08/12/1996. National Renewable Energy Laboratory. https://www.nrel.gov/docs/gen/fy08/42628.pdf
Sharma M, Soni R, Nazir A, Oberoi HS, Chadha BS (2011) Evaluation of glycosyl hydrolases in the secretome of Aspergillus fumigatus and saccharification of alkali-treated rice straw. Appl Biochem Biotechnol 163:577–591. https://doi.org/10.1007/s12010-010-9064-3
Peterson R, Nevalainen H (2012) Trichoderma reesei RUT-C30 - Thirty years of strain improvement. Microbiology 158:58–68. https://doi.org/10.1099/mic.0.054031-0
Dwiarti L, Boonchird C, Harashima S, Park EY (2012) Simultaneous saccharification and fermentation of paper sludge without pretreatment using cellulase from Acremonium cellulolyticus and thermotolerant Saccharomyces cerevisiae. Biomass Bioenergy 42:114–122. https://doi.org/10.1016/j.biombioe.2012.02.019
Liu G, Qin Y, Hu Y, Gao M, Peng S, Qu Y (2013) An endo-1,4-β-glucanase PdCel5C from cellulolytic fungus Penicillium decumbens with distinctive domain composition and hydrolysis product profile. Enzym Microb Technol 52:190–195. https://doi.org/10.1016/j.enzmictec.2012.12.009
Solís S, Loeza J, Segura G, Tello J, Reyes N, Lappe P, Guitérrez L, Ríos F, Huitrón C (2009) Hydrolysis of orange peel by a pectin lyase-overproducing hybrid obtained by protoplast fusion between mutant pectinolytic Aspergillus flavipes and Aspergillus niveus CH-Y-1043. Enzym Microb Technol 44:123–128. https://doi.org/10.1016/j.enzmictec.2008.11.003
Gunashree BS, Venkateswaran G (2010) Enhanced phytase production through interspecific protoplast fusion of Aspergillus niger CFR 335 and Aspergillus ficuum SGA 01 auxotrophic mutants. Enzym Microb Technol 45:562–567. https://doi.org/10.1016/j.enzmictec.2010.03.006
Ogawa K, Ohara H, Toyama N (1988) Intraspecific Hybridization of Aspergillus awamori var. Kawachi by Protoplast Fusion. Agric Biol Chem 52:337–342. https://doi.org/10.1271/bbb1961.52.337
Ogawa K, Tsuchimochi M, Taniguchi K, Nakatsu S (1989) Interspecific Hybridization of Aspergillusus amiimut. shirousamii and Aspergillus niger by Protoplast Fusion. Agric Biol Chem 53:2873–2880. https://doi.org/10.1271/bbb1961.53.2873
Wang H, Zhai L, Geng A (2020) Enhanced cellulase and reducing sugar production by a new mutant strain Trichoderma harzianum EUA20. J Biosci Bioeng 129:242–249. https://doi.org/10.1016/j.jbiosc.2019.08.016
Papzan Z, Kowsari M, Javan-Nikkhah M et al (2020) Strain Improvement of Trichoderma spp. Through Two Steps Protoplast Fusion for Cellulase Production Enhancement. Can J Microbiol. Advance online publication. https://doi.org/10.1139/cjm-2020-0438
Sabatier A, Neville-Marshall F, Nigel-Paterson H (2000) Enzyme Mixtures. Patent Application EP98401101A.
Liu F, Wang Z, Manglekar RR, Geng A (2019) Enhanced cellulase production through random mutagenesis of Talaromyces pinophilus OPC4-1 and fermentation optimization. Process Biochem 90:12–22. https://doi.org/10.1016/j.procbio.2019.11.025
Perkins, JB, Kumar M, Van Zijl MFK, Vollebregt AWH, Sarantinopoulos P (2012) DSM IP Assets BV. Talaromyces strains and enzyme compositions. U.S. Patent Application 13/381,871.
Li Y, Liu C, Bai F, Zhao X (2016) Overproduction of cellulase by Trichoderma reesei RUT C30 through batch-feeding of synthesized low-cost sugar mixture. Bioresour Technol 216:503–510. https://doi.org/10.1016/j.biortech.2016.05.108
Jourdier E, Poughon L, Larroche C, Monot F, Chaabane FB (2012) A new stoichiometric miniaturization strategy for screening of industrial microbial strains: application to cellulase hyper-producing Trichoderma reesei strains. Microb Cell Factories 11:70. https://doi.org/10.1186/1475-2859-11-70
Kubicek CP, Messner R, Gruber F, Mandels M, Kubicek-Pranz EM (1993) Triggering of cellulase biosynthesis by cellulose in Trichoderma reesei. Involvement of a constitutive, sophorose-inducible, glucose-inhibited β- diglucoside permease. J Biol Chem 268:19364–19368
Ilmén M, Thrane C, Penttilä M (1996) The glucose repressor gene cre1 of Trichoderma: Isolation and expression of a full length and a truncated mutant form. Mol Gen Genet 251:451–460. https://doi.org/10.1007/s004380050189
Le Crom S, Schackwitz W, Pennacchio L, Magnuson JK, Culley DE et al (2009) Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. Proc Natl Acad Sci U S A 106:16151–16156. https://doi.org/10.1073/pnas.0905848106
Nakari-Setälä T, Paloheimo M, Kallio J et al (2009) Genetic modification of carbon catabolite repression in Trichoderma reesei for improved protein production. Appl Environ Microbiol 75:4853–4860. https://doi.org/10.1128/AEM.00282-09
Orejas M, MacCabe AP, Pérez González JA et al (1999) Carbon catabolite repression of the Aspergillus nidulansxlnA gene. Mol Microbiol 31:177–184. https://doi.org/10.1046/j.1365-2958.1999.01157.x
Tamayo EN, Villanueva A, Hasper AA, Graaff LH, Ramón D, Orejas M (2008) CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet Biol 45:984–993. https://doi.org/10.1016/j.fgb.2008.03.002
Le Guilloux V, Schmidtke P, Tuffery P (2009) Fpocket: An open source platform for ligand pocket detection. BMC Bioinform 10:168. https://doi.org/10.1186/1471-2105-10-168
Jendele L, Krivak R, Skoda P, Novotny M, Hoksza D (2019) PrankWeb: web server for ligand binding-site prediction and visualization. BioRxiv 590877. https://doi.org/10.1101/590877
Magliery TJ, Regan L (2005) Sequence variation in ligand binding sites in proteins. BMC Bioinform 6:240. https://doi.org/10.1186/1471-2105-6-240
Guharoy M, Chakrabarti P (2005) Conservation and relative importance of residues across protein-protein interfaces. Proc Natl Acad Sci U S A 102:15447–15452. https://doi.org/10.1073/pnas.0505425102
Mintseris J, Weng Z (2005) Structure, function, and evolution of transient and obligate protein-protein interactions. Proc Natl Acad Sci U S A 102:10930–10935. https://doi.org/10.1073/pnas.0502667102
Schueler-Furman O, Baker D (2003) Conserved residue clustering and protein structure prediction. Proteins Struct Funct Genet 52:225–235. https://doi.org/10.1002/prot.10365
Matsumoto H, Koganei K, Nishida N, Koyama Y, Saito S, Kataoka H, Ogihara J, Kasumi T (2014) Cell dispersion culture for the effective growth of Humicola insolens and efficient enzyme production. J Biosci Bioeng 117:257–262. https://doi.org/10.1016/j.jbiosc.2013.08.014
Mahajan C, Basotra N, Singh S, di Falco M, Tsang A, Chadha BS (2016) Malbranchea cinnamomea: A thermophilic fungal source of catalytically efficient lignocellulolytic glycosyl hydrolases and metal dependent enzymes. Bioresour Technol 200:55–63. https://doi.org/10.1016/j.biortech.2015.09.113
Berrin JG, Navarro D, Couturier M, Olivé C, Grisel S, Haon M, Taussac S, Lechat C, Courtecuisse R, Favel A, Coutinho PM, Lesage-Meessen L (2012) Exploring the natural fungal biodiversity of tropical and temperate forests toward improvement of biomass conversion. Appl Environ Microbiol 78:6483–6490. https://doi.org/10.1128/AEM.01651-12
Acknowledgement
The financial support from the Department of Biotechnology, India, for carrying out research project “Developing fungal strains for enzyme production through mutagenesis, protoplast fusion and over expression using A. niger citrate synthase promoter” (BT/PR15271/PBD/26/509/2015) and from the Department of Science and Technology, India, for carrying out research project “Manipulation of transcriptional factors for developing hypercellulase producing strains for bioconversion of lignocellulosic biomass” (SR/WOS-A/LS-260/2018) is duly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 413 kb)
Rights and permissions
About this article
Cite this article
Basotra, N., Kaur, B., Raheja, Y. et al. Developing and evaluating lignocellulolytic hyper producing deregulated strains of Mycothermus thermophilus for hydrolysis of lignocellulosics. Biomass Conv. Bioref. 13, 5059–5071 (2023). https://doi.org/10.1007/s13399-021-01539-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13399-021-01539-1