Abstract
With advances in genetic research, the understanding of the genetic structure of disease and the ability to predict disease risk have been enhanced. Polygenic risk scores (PRS) have been developed to assess a person’s risk of developing any heritable disease. PRS has two primary utilities that make it particularly relevant for insurers: the ability to identify high-risk groups when using PRS independently or in combination with standard risk factors; and the ability to inform early interventions that may alter future morbidity and mortality. Using heart disease as a case study, a simulation-based model is designed that introduces polygenic risk scoring into the actuarial analysis framework and then quantifies the adverse selection due to information asymmetry introduced by PRS. Individual and parental disease liability as well as PRS were simulated under a liability threshold model. A series of validations were conducted to confirm the utility of our simulated data sets. We explored three scenarios describing how insurance applicants use their PRS results to guide their insurance purchasing decisions and calculated the increased premiums that insurers would need to change to counteract this. The accuracy of PRS has the most significant impact on premiums and the proportion of individuals who know their PRS also has a substantial impact.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
1.1 Genetic information in life and health insurance
In life and health insurance underwriting, insurers build actuarial models using various risk factors to estimate the likelihood of future health-related events, like disease and death, and to group individuals with similar risk profiles into homogeneous risk classes. Having access to all relevant information enables life insurers to fairly assess and price risk in the interest of all of their customers. However, in practice, insurers have many considerations and even restrictions on what risk factors they can use in insurance risk classification systems [7].
Genetics plays an important role in understanding human diseases. With the deepening of human understanding of the mechanisms of disease, the role of genetics in the process of disease has become clearer. Martin et al. [40] summarized two major roles of genetics on the understanding of human diseases “as a transformative line of etiological inquiry and as a biomarker for heritable diseases”. Statistical genomics studies have shown that common complex disorders have a polygenic genetic architecture, and have been able to identify genetic variants associated with diseases of interest. Today, integrating genetic risk information and non-genetic factors, the probability of individuals developing or experiencing disease over a specified time can be estimated using absolute or relative disease risk models [44]. By combining the list of disease associated genetic variants with their effect sizes, polygenic risk scores have been developed in recent years to represent genetic risk information.
The insurance industry’s use of genetic information is a complex issue that invokes extensive debate, including moral considerations, progress in empirical research, and balancing interests among various stakeholders. This controversy has lasted for more than 30 years, since the Human Genome Project began (October 1st 1990 [24]). If insurance companies are allowed to use genetic information, the risk of genetic discrimination occurs. Many people are worried that their genetic information may be misused by insurers, which might have an effect on the available insurance options for them and their family [2]. Moreover, because genetic information is bestowed on everyone at birth, many critics believe that it is unjust for health insurers to use such information [13]. Additionally, whether insurance companies would use genetic information properly is also controversial. The premise that insurance companies can use genetic information “well” is that genetic information can provide accurate information about disease risks. Opponents worry that insurance companies may, accidentally or otherwise, misuse genetic information, as knowledge of the genetic basis of disease is still incomplete. Finally, since family history as an underwriting risk factor can provide information about the inheritance of diseases between families, whether or not genetic information can provide sufficient additional useful information is still in question.
However, the sustainability and profitability of the insurance industry can be threatened by restricted access to risk predictors. Adverse selection cost is the best known threat, which comes from the asymmetry of access to predictive information between insurers and the insured. Today, individuals can easily obtain genetic information that impacts their future health status from direct-to-consumer (DTC) tests. Once customers have predictive information for common disease genetic risk, they can make a judgment about their future disease predisposition. It is then reasonable to assume that customers will take the above personal judgment into account when purchasing insurance. The simplest assumption is that people with a genetic report showing high risk are more likely to purchase insurance and those with low risk are less likely to purchase insurance, but insurers cannot distinguish between these high-risk and low-risk groups. As the foundation of insurance is that the majority of healthy people subsidize the minority of vulnerable people, this information asymmetry can be damaging, such as in cases where it leads to the insurer increasing premiums for all policyholders to compensate for an increased risk profile, including policyholders who are individually low risk. Note that we consider adverse selection through the prism of increased/decreased probability of purchasing insurance but do not investigate the possibility of increased likelihood of individuals buying abnormally large amounts of insurance, the latter of which is suggested to be the most expensive part of adverse selection in some studies [20].
After many years of discussion and research, most Western countries now hold a negative attitude towards whether genetic information can be used by insurance companies. Many countries have issued government regulations or industry self-regulation to restrict the access to genetic information by life insurers. Bélisle-Pipon et al. [2] summarized major developed countries’ considerations and latest regulation approaches towards insurer access and use of genetic information. Regulations will always change as the environment changes. Nevertheless, insurance regulators and insurers maintain a consensus that insurance practitioners should pay attention to the progress of scientists in genetic research and assess the impact of these developments on insurance [26]. Prince [49] provides a good recent account of attitudes towards genetics and insurance companies.
1.2 Insurance and genetics research literature review
When it comes to quantitatively measuring the impact of genetics on insurance participants, two main issues should be properly addressed: how to introduce genetic information into the actuarial framework and how to measure the impact of genetic information once such information has been successfully added to the actuarial framework.
Regarding the introduction of genetic information into the actuarial framework, quantitative research in this area is constantly updated with the discovery of disease-associated genes in genetic studies. Early on, genetic studies detected strong associations between specific genes and the risk of hereditary diseases, and such associations can also be verified in the biological pathway. For example, the risk of adult polycystic kidney disease (APKD) has been linked to the onset of APKD1 and APKD2 mutations. Mutations in the Huntingtin’s disease (HD) gene predominantly determine the onset of HD and inherited mutations in the BRCA1 and BRCA2 genes can lead to the onset of breast cancer. MacDonald et al conducted a series of studies to estimate the impact of disease related genetic information on insurance (e.g. [16, 17, 19, 33,34,35]). It should be noted that papers including [16, 17, 19, 33,34,35] make generic conclusions that genetic links are unlikely to be financially significant, which is something we attempt to quantify in this paper. Their studies estimated the probabilities of disease onset for each mutation at all age ranges under different circumstances, such as having or not having access to family history, based on epidemiological data regarding the onset of related diseases and other risk factors. The numbers of mutations involved in those studies are finite and are usually small. Those probabilities were then used to calculate transition intensities in multi-state Markov models, which were used to model the possible states involved in the policy time of an insurance customer. Under the Markov models framework, premiums of different insurance products for policy holders at any age of interest for a given period can be calculated. The cost of adverse selection, which is caused by the information asymmetry between insurers who do not have access to genetic information and the policy holders who have genetic testing results available, can be measured under reasonable assumptions about customer behaviour and the size of insurance market. For the interested reader, Chatterjee et al. [4] provide a thorough discussion of issues underlying adverse selection and conclude that under certain realistic assumptions, social welfare and insurance loss coverage can be increased by a ban on insurance risk classification
A far larger number of genes have been found to be associated with the onset of common disorders, like coronary artery disease, and simulation based research has begun to explore the impact of such multifactorial genetic disorders on insurance. Macdonald et al. [36] simulated a large-scale data set to estimate age-specific odds ratios for a 2 \(\times \) 2 gene-environment interaction model, which were used to parameterise a model of critical illness insurance. Macdonald and Tapadar [37] used the same 2 \(\times \) 2 gene-environment interaction model to explore the economic impact of multifactorial genetic disorders on critical illness insurance.
Regarding how to measure the impact of genetic information, comparing premiums across limited numbers of genotypes and measuring the cost of adverse selection using well-defined indicators are common strategies. Usually premiums were calculated under a range of insurance settings to give intuitive comparisons and adverse selection was monitored, capturing the information asymmetry around customers’ genetic information between consumers and insurers [19, 34, 35]. Sensitivity analyses were commonly employed to give a possible range for the cost of adverse selection. Generally, changing the parameters involved in the calculation of transition intensities also changes the severity of adverse selection in the sensitivity analysis, for example the mutation frequencies, penetrances and the onset of disease from epidemiological study. Therefore, the cost of adverse selection varies across different situations. Howard [20, 21] used the increased critical illness claim rates to represent the cost of adverse selection under the assumptions that insurers were banned from having access to genetic information and customers who tested positive tended to buy extra insurance. Howard [20, 21] concluded that as more and more people undergo genetic testing, this risk would increase and threaten the future stability of the insurance industry.
1.3 Today: polygenic risk scores in insurance
With understanding of the genetic basis of human disease deepening [6], a consensus has been reached that most human diseases with high incidences, like asthma, diabetes and cancers, are multifactorial disorders, which are affected by multiple genes and by interactions with environmental causes. Single nucleotide polymorphisms (SNPs) are the most common types of genetic variants and SNP array data is the widely used type of human genome data to detect the associated variants for the disease of interest. Such association studies carried out on various research cohorts have found that thousands of SNPs are statistically associated with the binary disease phenotypes, with each SNP only having a modest impact on the risk of each disease. Polygenic risk scores (PRS), also called genetic risk scores, are created by combining effects of a large set of statistically significant variants into a single score to represent the individual-level genetic risk of the disease under study. Section 2.2.4 gives more technical details on the calculation of PRS.
As an individual level genetic risk indicator, two major utilities of PRS make it especially important for insurers: one is the ability to identify high-risk groups when PRS is used independently or jointly with other risk factors; another is the ability to inform early interventions, which may change the future morbidity and mortality. The proposed use of PRS emerged with the development of statistical genomics research and whole-genome sequencing technology in recent years. Since the International Schizophrenia Consortium [50] reported that PRSs calculated from 37,655 SNPs on schizophrenia had the ability to predict up to 3% of the liability in independent case–control samples in 2009, PRSs for many common diseases have been developed and have demonstrated potential for disease onset risk prediction. Plomin and Von Stumm [46] summarized that there have been 2783 publications calculating PRSs on common human disorders and complex traits since 2009, with the predictive power of PRS increasing during the past decade. The clinical usage of PRS has been discussed widely by academia because adding PRS to the existing combination of clinical risk predictors has been shown to improve the accuracy of risk stratification for heart disease and other illnesses [63]. Informing PRS-identified high-risk subgroups of the population of their lifetime risk and then encouraging them to consider prevention strategies has the potential to reduce their risk of disease. A study conducted by Khera et al. [27] found that genetic risk can be attenuated by a favourable lifestyle among study participants at high genetic risk of coronary artery disease. The follow-up study carried out by Widén et al. [62] found that about half of individuals at high 10-year atherosclerotic cardiovascular disease risk had made health behaviour changes, including seeing a doctor, losing weight, quitting smoking, and signing up for health coaching online.
Not only is the use of PRS growing rapidly in academia, PRS is also getting closer to everyday lives. For example, as a leading company providing direct-to-customer (DTC) genetic testing services, 23andMe started to provide their customers’ risk of developing Type 2 diabetes based on their genetic profile, in 2019. Folkersen et al. [12] built a non-profit genetics analysis site, called https://www.impute.me/, to provide PRS calculations and results interpretations for DTC customers by allowing them to simply upload their genetics data obtained from DTC vendors. https://mygenerank.scripps.edu/ is a mobile app for coronary artery disease risk calculation and explanation, which allows 23andMe adult customers to get their polygenic scores after uploading their genetic data. GenoPred [45] is another tool to translate PRS into easy-to-understand results.
Insurance participants have been aware of the potential threat brought about by PRS in recent years. Vukcevic and Chen [60] introduced PRS as the genetic risk prediction indicator for complex traits and examined the impact of PRS-based testing on major diseases leading to critical illness claims, including coronary artery disease (CAD), breast cancer and prostate cancer. For every disease, they assumed there were only two types of risk related to it based on each person’s PRS level: ‘high risk’ and ‘low risk’. For example, they assumed that there were 20% of individuals who belonged to high risk for CAD and there existed 45% increase in risk relative to the remaining 80% ‘low risk’ group. Other insurance purchasing behaviour related assumptions were also made to quantify the impact of PRS on insurers, like the proportion of the population that obtains PRS-based genetic tests and the proportion of in-force policies that lapse if known to have low genetic risk. Financial analysis and sensitivity analysis were conducted with the focus on estimating the adverse selection impact of claim and lapse rates. The probability of an increase in claim cost, comparing the claims cost before anti-selection and after adverse selection, was estimated to measure the impact of adverse selection. Their results suggested that genetics was not a large threat because of the low proportion of the population who obtain PRS-based genetic tests, but adverse selection would become a threat when more people obtained their genetic tests. Swiss Re also warned of potential adverse selection risk brought by PRS on their website, including the accelerated loss developments from extra insurance cover sought by high risk groups and the need to develop new systems and methods to incorporate genetic data [54].
Reinsurance Group of America (RGA) and King’s College London carried out a study using real data on estimating the predictive power of PRS on common diseases and measuring the possibility of adverse selection brought by PRS [41]. Using genotype and phenotype data on UK Biobank participants, they confirmed the risk prediction ability of PRS on breast cancer (BC) and coronary artery disease (CAD) by comparing the hazard ratios (HRs) among different PRS risk groups. For example, the HR for the highest polygenic risk group had twice the risk of CAD compared with the middle risk group (HR = 1.97). They then quantified the impact of PRSs on adverse selection by assuming three different insurance purchasing behaviours with varying weight of PRSs on the decision of insurance purchasing. The cost of adverse selection brought about by PRSs rose with the increasing role of PRSs in the insurance purchasing decision. Lewis and Green [31] specifically mentioned that life insurers might use PRSs to adjust premiums, but must use and interpret PRSs carefully.
When measuring the impact of PRS on life insurers, the results PRS customers receive are the most direct indicators influencing their insurance purchasing behaviours. There are three main types of PRS results obtained from DTC genetic reports or online PRS calculation tools, which are the hazard ratios, the risk score position, and the absolute risk. Hazard ratios can be calculated from Cox regression models with the adjustment of other disease risk related parameters. This is the indicator used by Maxwell et al. [41]. One customer’s risk score position for the disease under study gives this customer’s quantile position on the overall population’s risk distribution. If one customer’s score lies at the \(15\%\) percentile, it means his/her genetic risk score for this disease is lower than \(85\%\) of and higher than \(15\%\) of the general population. This is one type of result obtained from https://www.impute.me/. One way to show absolute risk is by giving the incidence in the group of people with similar genetics to this customer and the incidence in the general population. Using the result from GenoPred [15], a person with PRS 2.36 (population prevalence 10%, polygenic score effect size type Cohen’s D, polygenic score effect size 0.7) has incidence of 33.2% among people with similar genetics, and incidence of \(10\%\) in the general population. Customers can be impacted by any of those risk indicators and then alter their insurance purchasing decisions.
1.4 Research goals
This study aims to introduce PRS for a common disease into the actuarial analysis framework through a simulation-based method. We use heart attack as a case–control study, as this disease has been well documented by Gutiérrez and Macdonald [16] and Tapadar [55]. We are aware that most real insurance policies cover multiple diseases. The simulation approach introduced in this study can be extended to any other disease of interest and the impact of PRS from each disease of interest can be analysed at the population level. The medical relationships between various diseases, such as comorbidities, are beyond the scope of this study. Insurance companies may not be able to get access to individual-level genetic information data resources, like the UK Biobank, or have access to customers’ genetic profiles, as most Western countries banned the usage of genetic information by insurers. Within the simulation setting, actuarial analysis can be carried out to measure the impact of PRS without direct access to real PRS data. Individual-level PRS is simulated for a large population to represent the genetic profile. Employing PRS as a continuous genetic risk gradient, the study population can be grouped into any number of subgroups of interest. In reality, the PRSs calculated on different research cohorts using different PRS methods have varying risk prediction ability. The accuracy of PRS is also considered in the simulation setup and the resulting differences are explored in various purchasing scenarios.
The simulation assumptions are explained in Sect. 2. We first simulate individual-level information for a large population, including age, sex, polygenic risk score, overall disease risk score, disease status, and family history status. The results for the validation of our simulated data sets and from the actuarial analysis are found in Sect. 3. Conclusions and further work follow in Sect. 4.
2 Methods
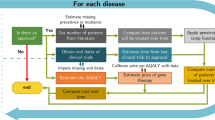
2.1 Assumptions for data set simulations
We first simulate individual-level information for a population of 500, 000 people, including age, sex, polygenic risk score, overall disease liability, their parents’ disease liability, and disease status of offspring and parents. Family history of cardiovascular disease has been well studied as an independent risk factor for coronary heart disease both in the short and long term, which identified the genetic contribution to disease susceptibility [1, 32]. All of these variables are sampled from their empirical real world distributions with appropriate stochastic relationships between variables. Disease related variables are related to each other and to non-disease variables according to established associations. Insurance purchasing behaviour is simulated under various adverse selection scenarios and then used to estimate the impact of PRS on insurers.
Simulated parental disease statuses are used only to obtain the family history of the offspring, while insurance purchase behavior is studied only for the offspring. Key assumptions are:
-
1.
All simulated individuals were assumed to belong to the same population, which in this study was the white British population. This is because, due to the complexity of the analysis, statistical genetics studies dedicated to establishing individual-level genetic risk are usually conducted in cohorts with the same ancestry, including the calculation of PRS [18].
-
2.
The age range was set from 0 to 69 years at age last birthday. The proportion of each age and sex group given in Table 1 was adjusted from the census data of the United Kingdom from 1995 [48]. The initial age for each individual was randomly uniformly simulated from the corresponding age range in that grouping. The age difference between parents and offspring is set to 30 years.
-
3.
Heart attack disease liabilities and PRSs for both parents and offspring were simulated under the liability threshold model (LTM). The LTM assumes that human disease risk can be measured by a hidden continuous liability, which is determined by the sum of inherited genetic liability (from parents) and the liability from the environment in which the offspring live. The threshold is a point on the continuous distribution of disease liability, which is determined by the prevalence of the disease in the sample population, and is used to distinguish cases and controls. The extent of inherited liability for different diseases varies and is measured by heritability. PRS is the genetic component liability that can be measured under current technology. Different PRS calculation methods capture varying degrees of inheritance liability, and thus the accuracy of PRS is embedded in the simulation. Section 2.2 explains this terminology in detail. See also Fig. 1.
-
4.
We assume that individual disease liability remains constant over the lifetime, but that thresholds vary with age. After simulating offspring age and sex, parent ages, offspring and parents’ overall disease liability and PRS, thresholds or prevalence rates are needed to determine the onset of heart attack. In this study, the age and sex-specific prevalence rates used in the LTM are derived from the heart attack transition intensities of a 4-state Markov model (see Fig. 2). The onset of parents’ heart attack is determined before the onset of their offspring. For parents, we assume that they have only two states, either a healthy state or a state where they have had a heart attack. The threshold point to determine every parent’s disease status is calculated using the independent lifetime heart attack probabilities. For offspring, we assume all start with a healthy state under this multi-state model and that any changes in state occur on a yearly basis, so we can count the number of heart attacks and deaths after each year’s simulation. Theoretically, the simulation can be conducted for any duration, but for the sake of simplicity we perform only 1 year of simulation in this study, and therefore we do not need to consider the newly added heart attack events for parents. We assume that the presence of family history is decided by the onset of the disease in either parent, as determined by the LTM. This may or not be the same as the definition of family history used by an insurance underwriter, but we believe it is a good proxy for family history even in cases where the definitions are not identical. Section 2.3 explains the strategy in detail.
-
5.
We assume the impact of PRS on insurers is influenced by the proportion of people who know their PRS results and the baseline percentage of simulated individuals who purchase insurance. Using such simulated data sets, insurers can estimate the severity of adverse selection brought about by PRS availability under various combinations of these two proportions. Section 2.4 provides a detailed description.
The liability threshold model (LTM) for a disease with prevalence K in the studied population. The unobserved continuous disease risk Y within the population is assumed to follow a normal distribution. Individuals with \(Y \ge t\) are those designated to have the disease of interest, otherwise they are designated to be healthy individuals
2.2 Individual-level variables simulation
This study employs concepts and models from human genetics to conduct our simulation. We start by explaining basic concepts in heritability of human diseases.
2.2.1 Basic concepts in heritability of human diseases
The onset and progression of human diseases are highly heterogeneous, but family aggregation of disease has been noticed and studied for a long time [10]. Heritability is the single widely used measure of the degree of resemblance between relatives on the onset of disease [56]. In quantitative genetics, many diseases are measured on a binary or 0/1 scale, with the absence of disease as 0 and the presence of disease as 1, and called phenotypes when used in statistical models. Complex disease heritability is defined as the total phenotypic variance in a population explained by the genetic component only, leaving the remaining unexplained variance attributable to environmental components or the interaction components between gene and environment as documented in the ensuing equation [11]:
where P is the phenotype, G is the genotype and E is the residual including anything other than genetics. Under this model, the total phenotypic variance (\(V_{P}\)) is partitioned into genotypic variance (\(V_{G}\)) and the environmental variance (\(V_{E}\)). The genotypic component can be partitioned further into additive genetic variance (\(V_{A}\)), dominance component variance (\(V_{D}\)), and interaction component variance (\(V_{I}\)):
Heritability is defined as the genetic variance \(V_{G}\) to the phenotypic variance \(V_{P}\), which ratio measures the degree to which an individual’s phenotypes are determined by their genotypes. Because the major cause of resemblance among relatives is the additive genetic variance (\(V_{A}\)), the ratio of the additive genetic variance to the phenotypic variance (\(V_{P}\)) is used to measure the degree to which an individual’s phenotypes are determined by the genes transmitted from their parents. This ratio is called narrow sense heritability, or simply the heritability,
2.2.2 Liability threshold model
The liability threshold model (LTM) assumes that there exists an underlying continuous variable to represent a person’s risk of getting a disease, which is the liability. When a person’s liability is above the threshold level the individual presents the disease; when the underlying variable is below the threshold the individual is classed as normal. The observed binary disease trait is transformed to the unobserved continuous liability scale under the liability threshold model. The liability threshold model was developed to estimate the inheritance of human disease by overcoming the “all-or-none” binary characteristic of the human disease phenotype [10].
Falconer [11] set the liability as being governed by a normal distribution in the LTM, a choice justified by the central limit theorem when the liability is constructed as the sum of a large number of independent terms, as per multifactorial common diseases. Letting t be the threshold value, individuals with disease liabilities Y greater than t are designated to have the affected phenotype and those with liabilities less than t are designated to have the normal phenotype. Figure 1 provides an illustration. The area to the right t is equal to the disease prevalence K, with \({\Phi }^{-1}(1-K) = t\), where \({\Phi }\) is the cumulative distribution function of the normal distribution.
2.2.3 Breakdown of disease liability
As explained in Sect. 2.2.1, the liability of disease Y can be assumed to the sum of additive genetic components \(Y_{g}\) and the combination of environment and unknown risk factors \(Y_{e}\). In accordance with the relevant literature [11], we also assume that \(Y_{g}\) and \(Y_{e}\) are independent and normally distributed with mean and variance \(\sigma ^{2}_{g}\) and \(\sigma ^{2}_{e}\). Falconer [11] explained that correlation between genotype and environment was an unimportant complication and could be ignored in experimental populations. Therefore the phenotype variance \(V_{Y}\) is taken to be the sum of \(\sigma ^{2}_{g}\) and \(\sigma ^{2}_{e}\). For the purpose of statistical analysis, it is common to set the mean values of \(Y_{g}\) and \(Y_{e}\) equal to 0 and the total phenotype variance equal to 1. Under those settings, the LTM can be written as:
The phenotypic variance explained by the genotype variance is:
which is exactly the definition of heritability in Eq. (2), so heritability equals the variance of the genetic component under the LTM framework, \(h^{2} = \sigma ^{2}_{g}\). Those settings are still valid in recent statistical genetics research (e.g. [23, 29, 52]). Heritability is a value between 0 and 1 for the disease of interest. The larger the ratio, the larger role genetics plays on the determination of that disease. Many factors have been found to play a role in the estimation of heritability for human diseases, including the type of disease, population structure and research design [56].
2.2.4 PRS calculations
The human genome is composed of a series of nucleic acid sequences, comprising approximately 3.2 billion nucleotides of DNA [3]. One of the primary goals of human genetics research is to identify which DNA sequence variants impact the onset and progression of human diseases. Sequencing the whole human genome is difficult. Instead, researchers commonly use genome-wide genetic variants sequencing, which sequences differences in individual DNA building blocks, called single nucleotide polymorphisms (SNPs). Statistical genomicists build statistical models to detect the associations between SNPs and diseases. Genome Wide Association Studies (GWAS) is the experimental design used to detect associations between SNPs and diseases in samples from populations with the purpose of better understanding the biology of disease [58]. GWAS usually restrains their study participants to belong to the same ethnic groups and subgroups to reduce the possibility of false-positive results [57].
Over 10,000 significant associations between SNPs and diseases have been detected so far [58], with most variants having only small effects on the development of diseases. Except for several genetic variants that have directly been established to translate from gene to biological influence on common diseases (e.g. BRCA1 and BRCA2 genes increasing the risk of female breast and ovarian cancers because both genes produce tumor suppressor proteins [51]), most variants have only small effects for the development of disease. This is because the SNPs either play a regulatory role or are simply correlated with the genes that directly impact disease biology.
Polygenic risk scores (PRS) were created by combining those small, but statistically significant, variant effects into a single PRS to measure the heritability of a trait and disease onset. PRS is calculated by the additive or multiplicative combination of a selected set of single nucleotide polymorphisms (target SNPs) and corresponding effect sizes from GWAS summary statistics into a single score to measure the heritability of a trait and disease onset. The additive PRS is calculated as the sum of independent risk variants multiplied by their corresponding effect sizes. Even though the additive PRS assumes an additive genetic architecture without modelling any gene-gene or gene-environment interactions, this architecture represents the current best estimate of genetic architecture of common multifactorial disorders [30]. The baseline function for additive PRS is
where X refers to the set of significant variants and \(x_{i}\) is the genotype at the \(i^{th}\) selected marker, such as single nucleotide polymorphisms (SNP) allele counts coded as 0, 1 and 2 for homozygous, heterozygous, and other homozygous genotypes. \(\beta _{i}\) is the log odds ratio or the effect size from GWAS summary statistics for binary and quantitative traits respectively.
PRSs can also be calculated through various algorithms with different performance. For example, PRSice [9] calculates several set of PRSs by including SNPs below different p-value thresholds and determines the best p-value threshold on the test data. Additionally, PRS can be constructed using modelled effect sizes to improve the explained heritability. Commonly used modelling methods are Beta shrinkage [38], Bayesian estimation [53] and linkage disequilibrium (LD) adjustment [59], or the combination of two of the methods [14]. Good references for better understanding on PRS can be found from [5, 46, 63].
2.2.5 PRSs in the liability threshold model
Since PRS summarizes the genetic components of risk into a single score, it can be used to represent individual-level genetic component risk. The central limit theorem indicates that the PRS of a large enough sample should approximately follow a normal (Gaussian) distribution [5], when selected genetic variants are independent from one another and all samples have the same ancestry. Therefore, the LTM can be written with PRS as the measured genetic component:
where \(PRS \sim N(0, \sigma ^{2}_{PRS})\), given \(Y \sim N(0, 1)\), so that the disease heritability explained by PRS equals the variance of PRS, \(h^{2}_{PRS} = \sigma ^{2}_{PRS}\), under this setting.
For the same disease of interest, assuming the hidden continuous liabilities are constant for a population, if there are two sets of PRS having different variances, the set with larger variance includes more genetic component information and leaves less uncovered genetic component information. For example, LDpred2 [47] calculates PRS on GWAS summary statistics and a correlation matrix between genetic variants. PRS derived by LDpred2 for coronary artery disease (CAD) explained about 8.5% of the phenotypic variance in the UK Biobank participants with the ROC (receiver operating characteristic) area under the curve (AUC) equal to 0.64 (see [22] for further details on ROC and AUC). Other PRS methods compared by Privé et al. [47] all had lower AUC values. The higher the predictive power of PRS, the higher the accuracy in differentiating between cases and controls, and the greater the potential impact of PRS on insurers. Research is ongoing into improving PRS estimation, therefore this study also explores the impact of PRS accuracy.
2.2.6 The liability resemblance between parents and offspring
Using \(Y_{p}\) and \(Y_{o}\) to represent the phenotypic liability for parents (either mother or father) and the phenotypic liability for offspring respectively, Falconer [11] pointed out that the resemblance between offspring and parents was expressed via the regression of offspring on parents. The liability relationship between \(Y_{p}\) and \(Y_{o}\) can be deduced as:
where \(cov(Y_{p}, Y_{o})\) is the liability covariance of offspring and one parent, and \(V(Y_{p})\) is the liability variance of one parent. Falconer proved that \(cov(Y_{p}, Y_{o}) = 1/2 V(A)\) (Falconer [11], Chapter 9), where V(A) is the additive genetic variance of the parents. Following this result, the liability relationship between \(Y_{p}\) and \(Y_{o}\) can be deduced as:
because heritability equals \(\frac{V_{A}}{V_{P}}\) (Eq. 2). Therefore
The liability covariance between offspring and one parent can be calculated by the heritability:
2.2.7 Simulating disease liabilities for parents and offspring jointly
We have derived the liability covariance between parent and offspring in Eq. (5). Combined with the liability threshold model assumptions in Eq. (3), we can write the covariance matrix for parent and offspring as:
Here, \(0.5h^{2}\) means that the resemblance in disease liability between one parent and offspring is half of the inherited genetic predisposition as each offspring inherits half of their risk from each parent. For offspring, when we use PRS to represent the measured genetic component, the covariance between offspring PRS and offspring disease liability is \(h^{2}_{PRS}\) (Eq. 4). Combining these relationships, disease liabilities for parents and offspring can be simulated jointly, with the relationship written as a covariance matrix for multivariate normal distributed random variables [23]:
where \(Y_{p1}\), \(Y_{p2}\) and \(Y_{o}\) are the hidden continuous liabilities of heart attack for parents and offspring and \(Y_{o,g}\) is the measured genetic component of the liability for offspring.
This study uses heart attack as a case study. Many factors can contribute to an individual’s first-ever heart attack. Coronary artery disease (CAD) is one major cause of heart attacks. In this study we use the estimated heritability for coronary artery disease to represent \(h^{2}\) for the first-ever heart attack in our simulation. CAD is a well-studied multifactorial genetic disorder, and heritability estimates for CAD range between \(40\%\) and \(60\%\) based on pedigree studies [42]. Current technology cannot locate all genes that have an effect on disease onset, so these values give an estimated upper bound of the phenotypic variance explained by the genetic component of CAD. In this study, we use 50% as the heritability of CAD, in other words, the total phenotypic variance explained by the entire genetic component is equal to 0.5. Therefore, the covariance between \(Y_{p}\) and \(Y_{o}\) is equal to \(0.5h^{2} = 0.5\times 0.5 = 0.25\). To represent realistic sets of PRS, we simulate PRS with different variances, \(h^{2}_{PRS}\) is set equal to 0.01, 0.1, and 0.3, all less than 0.5. Here, 0.01 represents the situation where PRS has almost no power to distinguish cases and controls. The second choice, 0.1, is close to the heritability explained by CAD-PRS calculated using the LDpred2 method [47]. The final value, 0.3, is the approximate upper limit of the variance explained by PRS across diseases [63]. This is because PRS only represents one type of genetic component (SNP), and there are other types of genetic information that contribute to the inheritance of disease.
2.3 Onset of heart attack for parents and offspring
The onset of heart attack for both parents and offspring is determined under the liability threshold model. After simulating offspring age and sex, parent ages, offspring and parents’ overall disease liability and PRS, thresholds or prevalence rates are needed to distinguish cases from controls. Prevalence rates come from a 4-state Markov model, which was used to model heart attacks by Tapadar [55]. We first simulate heart attack onset in the parents and then simulate the onset in the offspring.
2.3.1 The 4-state heart attack Markov model
This model is presented in Fig. 2, which assumes that every simulated participant starts at the healthy state, and then allows them to remain healthy or transition to the heart attack or dead (not from heart attack) states over the simulation period. Transition intensities are represented using the notation \(\mu _{x}^{12}\), \(\mu _{x}^{13}\), or \(\mu _{x}^{24}\) for the relevant pairs of connected states, where x denotes age.
The transition intensity between the healthy state and the heart attack state, \(\mu _{x}^{12}\), was first formulated by Gutiérrez and Macdonald [16] based on the numbers of first-ever cases of heart attacks between September 1991 and August 1992 taken from McCormick [43]. Table 2 replicates the 1-year incidence of first-ever heart attack formed by Gutiérrez and Macdonald [16], and they fit transition intensities for males and females separately. For males they fitted the following functions:
with linear interpolation between ages 44 and 49. For females they fitted
Transition intensities between healthy state and dead state, \(\mu _{x}^{13}\), measure the force of mortality affecting individuals who have not had a heart attack. We calculated all age transition intensities of non-heart attack deaths following the steps described by Tapadar [55], as in Fig. 3.
We are interested in comparing the group with heart attack after simulation to the overall population, therefore transition intensities between sickness state and dead state, \(\mu _{x}^{24}\) are not of interest. Hence, we didn’t consider the post-heart attack state (representing events like died within 28 days due to heart attack or died due to other causes after recovery from heart attack).
2.3.2 Heart attack events for parents
For the parents, after we simulated their age and disease liability, we assumed that they were alive and had only two states - with or without a heart attack - during their lifetime. We assume the probability of heart attack for parents is the independent life time heart attack probability:
where \(\mu _{x}^{12}\) is transition intensity for males (Eqs. 7 and 8) and females (Eq. 9) respectively. For example, for a female parent age 45, her probability of having heart attack is equal to \({}{_{45}q}_{0}^{12} = 1 - exp(-\int _{0}^{45}\mu _{45}^{12} du) = 0.001710967\) with \(\mu _{45}^{12} = 0.0003570431\) calculated from Eq. (9). Therefore each age and sex group has its own specific prevalence, and then the corresponding liability threshold can be calculated as \(t_{x} = {\Phi }^{-1}(1-{}{_{x}q}_{0}^{12})\). This is the threshold used to distinguish cases and controls. For populations of the same age and sex, individuals with above-threshold liabilities are designated as having heart attacks and with below-threshold liability are designated as having a healthy status. In this study, we only consider whether the parent has a heart attack from birth to their present age and assume they all alive at present. The year in which the heart attack occurred was not simulated and had no impact on our analysis, which is based on the offspring individuals only.
2.3.3 Heart attack events for offspring
For offspring, we assume all start with a healthy state under this multi-state model and that the changes in state are documented on a yearly basis. We perform 1 year of simulation in this study for simplicity. Since the heart attack state and the death from healthy state are competing events in this 4-state heart attack Markov model, dependent transitions probabilities must be calculated from the above independent transition probabilities in Eqs. (7), (8) and (9). Using transition intensities \(\mu _{x}^{12}\) and \(\mu _{x}^{13}\), the 1-year independent heart attack probability \(q_{x}^{12}\) and 1-year independent death probability \(q_{x}^{13}\) for integer age at last birthday were calculated:
because \(\mu _{x}^{12}\) is a continuous function for all age ranges and \(\mu _{x}^{13}\) is a point estimate function for each integer age.
Assuming that deaths and heart attacks occur on average halfway through the year, the 1-year dependent heart attack probabilities \(aq_{x}^{12}\) and the 1-year dependent death probabilities \(aq_{x}^{13}\) are calculated as:
Under the uniform distribution of deaths assumption, \({}{_{0.5}q}_{x}^{12} = 0.5 *q_{x}^{12}\) and \({}{_{0.5}q}_{x}^{13} = 0.5 *q_{x}^{13}\).
For each integer age and sex group, the corresponding 1-year liability threshold \(t_{x} = {\Phi }^{-1}(1-aq_{x}^{12})\) is assumed to come from the 1-year dependent heart attack probabilities \(aq_{x}^{12}\) and is used to determine the case and control status under the LTM for that age group. For example, we get \(aq_{55}^{12} = 0.00504\) and then \(t_{55} = {\Phi }^{-1}(1-aq_{55}^{12}) = 2.57\) for 55-year-old males. This threshold means that, during the 1 year period, 55 year-old males with disease liability greater than 2.57 transfer to heart attack state. Remaining males in the group are left for potential death state transfer selection. A random Bernoulli trial with the probability of \(aq_{55}^{13}\) is given to every remaining man in the group to check if this person has a random result of 1. If so, this man is moved to the dead state. Men, whose disease liability is less than the threshold and get random results 0 from Bernoulli trails, stay in the healthy state at the end of 1 year simulation.
2.4 Measuring the impact of PRS on insurers
If insurers and policyholders have access to the same information, there is no possibility of adverse selection. However, because insurers have been prohibited from getting access to customers’ genetic information, when insurance applicants know their PRS results and use that to guide their insurance purchasing behaviour, the potential for adverse selection arises. Following the simulation assumptions, we can obtain a large sample size of simulated data with individual-level information for offspring, heart attack status of their parents and the heart attack status of offspring after 1 year of simulation. We explore three scenarios (see Sect. 2.4.1) describing how insurance applicants use their PRS results to guide their insurance purchasing decisions.
When a greater number of individuals with high PRS values decide to buy insurance products, the proportion of heart attack events within the insured population at the end of the policy year is higher than the proportion of heart attack events in the overall simulation population. We measure the increased proportion of heart attack incidence among the insured group (as defined in Sect. 2.4.2) versus the overall simulated population for each scenario of interest. In the event that adverse selection happens, insurers can increase premiums to balance the increased payouts for more heart attack events among the insured population. Using the simulated data, insurers can calculate premiums for any groups of interest. In this study, we calculate premiums separately for the insured population and the overall population and measure the extent of the premium increase (see Sect. 2.4.3). Marketing considerations are beyond the scope of this study.
2.4.1 Adverse selection scenarios
The extent to which the general population possesses their genetic information and the response of insurance customers to knowing their PRS results are important indicators as to the potential severity of adverse selection. We record the proportion of simulated individuals who know their PRS results and the baseline probability of purchasing insurance for each simulated individual. In theory we can explore any scenario of interest, but we have selected three representative scenarios to reflect the overall trend of insurance applicants using their PRS results to purchase insurance. The first represents a high proportion of individuals knowing their PRS results and a relatively high intention to purchase insurance in the overall population, the second describes a high proportion of individuals knowing their PRS results but relatively low intention to purchase insurance in the general population, and the third describes a low proportion of individuals knowing their PRS results and a relatively low intention to purchase insurance in the overall population. The three scenarios considered are:
-
1.
100% of simulated individuals know their PRS results and 10% of individuals intend to purchase insurance as a baseline measure.
-
2.
100% of simulated individuals know their PRS results and 1% of individuals intend to purchase insurance as a baseline measure.
-
3.
10% of simulated individuals know their PRS results and 1% of individuals intend to purchase insurance as a baseline measure.
The choices for the scenario % values are somewhat arbitrary, but we simply wish to test representative values of low and higher insurance purchasing and examine any emerging trends. Interested parties will of course wish to adapt these settings to their own needs. It is also recognised that, for greater realism, insurance demand (% purchasing insurance) should ideally be a function of the premium charged, but we felt that this complicating factor was beyond the scope and focus of the paper.
Various measures can be used to represent a person’s genetic risk for a disease of interest, and hazard ratios (HRs) are one of the common results that individuals can get from today’s commercial genetic test reports to describe their disease risk. HRs calculated from Cox regression analysis were employed to explore the impact of PRS on adverse selection by Lewis and Vassos [30]. They evaluated the role of PRSs in assessing the risk of coronary artery disease and breast cancer using Cox regression analysis with age as the time-dependent variable and PRSs risk group as the explanatory covariate. The median PRS risk group, individuals with PRS within the 40–60% percentile range, is used as the reference group in their study and we replicate that approach here. The hazard function, \(\delta (t)\), is influenced by the PRS risk group (\(PRS_{group}\)). In this study, we follow the same PRS-based purchasing assumption used by Maxwell et al. [41], which assumes that increases in individuals purchasing insurance policies occurred as the hazard ratio for each PRS risk group versus the reference group increased. In other words, each PRS risk group buys insurance with probability proportional to its corresponding HR, taking into account the baseline insurance purchasing intention that is present.
2.4.2 Identify the insured group
This study assumes that every simulated individual has the same baseline proportion of purchasing insurance until they know their PRS results. Either 100% or 10% of individuals (based on which scenario is under consideration) are randomly simulated as possessing their PRS results. Then, every simulated individual’s probability of insurance purchasing is adjusted based on whether they know their HR values as a result of possessing their PRS results or not. Finally, random Bernoulli trials determine the subset of individuals who do buy insurance. The insured group is determined under each scenario of interest.
-
1.
Using all simulated data, calculate HRs for each PRS risk group using the median risk group as the reference group.
-
2.
Randomly select individuals as having the status of knowing their PRS results, where PRS results are captured in the form of HRs.
-
3.
For individuals who know their HRs, adjust their probability of purchasing insurance. For example, if the HR is 2 for the \(90{-}100\%\) PRS risk group, under scenario 1, the probability of purchasing insurance for this high risk group is \(2 \times 10\% = 20\%\). When the HR-based proportion is greater than \(100\%\), we use \(100\%\).
-
4.
For every simulated individual, determine if they purchase insurance using the Bernoulli distribution with the input parameter as their probability of purchasing insurance. For individuals who know their HRs, they have adjusted probabilities as their inputs, and for individuals do not know their HRs, they have the baseline probability of purchasing insurance.
-
5.
For each simulated data set, steps 2 to 4 are repeated 100 times to counteract simulation basis.
2.4.3 Premiums calculation
Insurance premiums for policyholders measure the cost of future possible payouts by insurers, with high-risk customers paying a larger premium. Following the principle of equivalence, premiums can be calculated under various policy settings. For example, Gutiérrez and Macdonald [16] and Macdonald et al. [33] used transition intensities between any two connected states from a multi-state Markov model to calculate the continuous payable premiums for any given age with a given policy term on the insurance contract of interest. Their transition intensity functions were calculated and smoothed from grouped incidence data recorded in medical studies. However, smoothing transition intensity functions is not the focus of our study. We employ a straightforward single critical illness policy, which assumes payment of a lump sum of \(\pounds 10{,}000\) is made at the end of the policy year if the policyholder has a heart attack during the policy year. We calculate premiums for a 1-year standalone policy, using group heart attack incidence rates as an approximation of the transition intensity between the healthy state and the heart attack state in the 4-state heart attack Markov model (Fig. 2). Premiums are calculated as the expected present value (EPV) for such a payment with an assured \(4\%\) interest rate per year:
where \({}{_{1}q}_{x}^{12}\) is calculated using Eq. (10).
3 Results
To validate the ability of our simulated datasets to mimic real data, we check the ability of PRS to distinguish cases and controls and compare the incidence rates in each sex and group in our simulated dataset with the incidence rates among real-world subjects. The results of the comparison are satisfactory. The impact of PRS accuracy is visualized by comparing the proportion of heart attack events for males and females under different PRS variance settings and comparing the cumulative incidence rates of heart attack. The risk stratification ability of family history and PRS is compared and assessed via the change in incidence rates. The simulated dataset is then used for actuarial analysis with the aim of quantifying the impact of PRS on insurers. Grouped premium calculations, Cox regression analysis and adverse selection measures are the three aspects of actuarial analysis involved in this study. In this study, we set \(h^{2}_{PRS}\) equal to 0.01, 0.1, and 0.3. To avoid simulation bias, 100 simulation replications were produced for each value of \(h^{2}_{PRS}\) and means and confidence intervals were subsequently calculated.
3.1 Simulation data sets validation
3.1.1 Summary statistics
A variety of individual-level information is simulated, including age, sex, polygenic risk score (PRS), overall disease liability, disease liability of parents and disease status of offspring and parents. Table 3 lists the summary statistics of the continuous variables after 1 year of simulation from a single set of simulated data, with the variance of the simulated PRS set to 0.3. To avoid missing any elements, we have removed the columns for those variables and retained only the quantitative explanation. The variance of PRS also equals the heritability of PRS under the LTM in this study. The starting age covers 0 to 69 at age last birthday (1). The age difference between both parents and offspring is 30 years (Assumption 2 in Sect. 2.1). The simulation of offspring PRS, offspring disease liability and parental liability employs the multivariate covariance matrix in Eq. (6).
Parental and offspring heart attack status was determined by the lifetime independent probability of heart attack and 1-year dependent probability of heart attack as calculated in Sect. 2.3. Table 4 shows the proportions of each event calculated from the same data set. The simulation period is only 1 year, so the heart attack incidence rates and the death rate are quite low. The earliest age at the last birthday of a simulated offspring heart attack onset is 16 years. All other simulated data sets under the same settings have similar outcomes.
In this study, we assume that the presence of family history is determined by the onset of the disease in either parent. This is not the same as the definition used by insurance underwriters but is a reasonable proxy. For example, there are in total 64, 581, 30, 011, and 603 heart attack cases for father, mother and offspring respectively calculated from the same simulated data set used in Tables 3 and 4. There are 8602 offspring that have both parents having experienced heart attacks. Following our assumptions, about 17% of offspring have a family history of heart attack. Amit Khera, M.D. from the Department of Internal Medicine at UT Southwestern Medical Center, estimated that \(10{-}15\)% of the U.S. population has a strong history of heart disease [61], so this outcome seems reasonable. In our simulation, we simulated age distribution following the censored data distribution of the United Kingdom from 1995.
3.1.2 PRS distribution
Polygenic risk scores are generated for the overall 500,000 simulated population following a normal distribution with a mean of 0 and a variance of \(h^{2}_{PRS}\). Figure 4 presents the cases and controls violin plots for PRS and the polygenic score percentile plots, which are generated from the same simulation dataset used to create the above summary statistics table and events table. The grey violin shape represents the PRS density for cases, and the blue violin shape is for controls. The box elements within both violins indicate that the median PRS for cases are higher than controls, and there is a significant difference between the density plots of cases and controls. The reason for the volatility in the cases violin plot is due to the relatively low heart attack incidence, only accounting for 0.126% of the total population. The median value for controls is centred around 0, because controls account for the majority of simulated data. The overlap between cases and controls indicates that PRS does not have 100% ability to distinguish cases and controls.
Figure 4b shows the prevalence of heart attacks in the 100 groups binned according to the percentile of PRS, with the x-axis representing the 100 polygenic score percentile groups, arranged from the lowest PRS value to the highest PRS value. The variance of PRS is equal to 0.3. There are few heart attack events for the first 75 PRS percentile groups, so prevalences for those groups are close to zero. Thereafter, the prevalence increases with increasing PRS values as expected, so we see more heart attack events in the high PRS risk groups than in the low PRS risk groups. We also expect to see some heart attack events occurring in the relatively low PRS risk groups, as PRS does not explain all disease risk. These two graphs show results that are consistent with our expectations. The shape of Fig. 4 is similar to that of Figure 2 from Khera et al. [28], which shows the risk of CAD according to PRS calculated on UK Biobank participants.
3.1.3 Grouped heart attack incidence
We compare the age and sex specified incidence calculated from our simulated datasets with the grouped incidence in Table 2. Following the same simulation setting with a sample size of 500,000 and a PRS variance of 0.3, 100 simulation replicates are produced. The grouped heart attack incidence rates are calculated from all simulated datasets and then used to calculate the mean rates and the corresponding confidence intervals for the incidence rate of that group. Figure 5 shows the mean and 95% confidence interval of incidence rates for both females and males of all 7 age groups ranked from youngest to oldest. The top and bottom of each error bar represents the 0.975 quantile and 0.025 quantile of that group’s incidence rates. The solid square points on each error bar indicate the position of that group’s real incidence in Table 2. Those real incidences were calculated using numbers of first-ever cases of heart attacks between September 1991 and August 1992, taken from the Morbidity Statistics from General Practice Survey [43].
The incidence rates calculated from our simulated datasets show a similar trend to the empirical incidence rates, i.e., the incidence rates are always higher in males than in females and the incidence rates increase with age. Figure 5 shows that the solid square points always lie within the confidence intervals, most of which are close to the corresponding mean locations. Males within the age group [60, 64] is an outlier because the actual incidence in this group lies at the far end of the error bar. One possible explanation is that the observed incidence for this group might not represent the truth in the longer term. The risk of contracting the disease should increase with age, but the actual incidence in this age group from table 2 is very close to the value of the nearest younger group, i.e., those aged between [55, 59] on their last birthday. In general, our simulated incidence rates are very close to the morbidity statistics values.
Grouped heart attack incidence calculated from 100 simulation replicates, with each simulation having sample size of 500, 000 and heritability \(h^2_{HA}\) of 0.30. The top and bottom of the error bars represent the 0.975 quantile and 0.025 quantile of corresponding group’s incidence rates. Those solid square points on each error bar indicate the position of the original incidence from Table 2
3.1.4 Risk stratification ability by family history and PRS
The disease statuses for parents and offspring are explained in Sect. 2.3. Figure 6 shows the age-grouped incidence for groups with and without family history and for three PRS risk groups separately. As expected, family history groups have higher incidences than groups without family history for both males and females at all 7 age groups. Bottom two plots show the incidence for three different PRS risk groups separated by their PRS values. The highest PRS risk groups have significantly larger incidences for both males and females at all age groups. Both medium and low PRS risk groups have zero incidence due to the rare heart attack events in the simulated data sets. Comparing female plots only, incidence rates for the ‘with family history’ group are between the incidences for the PRS \(40{-}60\)% and \(80{-}100\)% groups. This suggests that PRS and family history provide overlapping information. We can draw simple conclusions from this, such as if an insurance applicant has a family history of heart attack, he/she has a higher probability of belonging to a high PRS risk groups than a without family history applicant. This finding also holds for males.
Heart attack incidence for a groups with and without family history and for b groups with various PRS risks. Points used to create those plots are the mean incidences and the corresponding 0.975 quantile and 0.025 quantile values from 100 simulation replicates, with each run having sample size 500,000 and the phenotypic variance explained by PRS (\(h^2_{PRS}\)) equal to 0.30. Note the larger error bars in a, which means that using family history as a risk factor introduces more uncertainty than using PRS
Heart attack incidence rates for PRS risk subgroups with and without family history (FH). Points used to create those plots are the mean incidence and the corresponding 0.975 quantile and 0.025 quantile values from 100 simulation replicates, with each run having sample size 500,000 and the corresponding heritability
Figure 7 shows the grouped incidence for family history and PRS jointly. To create this plot, we first split the simulated dataset into two groups: with family history and without family history. Then within each group, we calculate the age and sex grouped incidence for three different PRS risk subgroups. Therefore, there are in total 6 risk groups for both males and females. Again, points used to create Fig. 7 are also the mean incidence and the corresponding 0.975 quantile and 0.025 quantile values from 100 simulation replicates. Solid and dashed lines are used for groups with family history and without family history respectively. Figure 7a presents the results when \(h^{2}_{PRS}=0.3\). For both males and females, only the high PRS risk groups with and without family history have positive incidence rates; all other subgroups have zero incidence rates at most age groups. The high PRS risk subgroup with family history always has higher incidence than the subgroup without family history. The low proportion of heart attack events is the reason for the zero incidences in the simulations. Figure 7b shows the results when \(h^{2}_{PRS}=0.1\). In the real-world cases, as the PRS variance decreases, more heart attack events occur in the middle and low PRS risk groups, although women and men in the high PRS risk group still have the highest incidence. It’s worth noting that the middle PRS risk group (PRS quantile 40–60%) with family history has similar incidence to the high PRS risk group (PRS quantile 80–100%) without family history, even though incidence values are slightly different between females and males.
Results shown in Fig. 7 are consistent with the study of Mars et al. [39], which states that family history and PRS are independent measures but can provide complementary information on susceptibility to most inherited diseases, including coronary artery disease. Mars et al. [39] pointed out that a positive family history with high PRS is associated with a fairly high risk, while low PRS completely compensates for the risk implied by a positive family history.
3.2 Visualization of PRS accuracy
Figure 8 visualizes the effect of different sizes of phenotypic variants explained by PRS on the proportion of heart attack events in men and women. The y-axis gives the percentages of the number of heart attack events in each PRS strata versus the total number heart attack events from the overall dataset. Therefore, for every single line on those plots, the sum of all five PRS groups’ percentages are all equal to 100%. Percentages used to create those plots are all mean percentages calculated from 100 simulation replicates under the same settings. When the value of PRS-based heritability is low, PRS only plays a small part of the determination of disease onset and its ability to distinguish cases and controls is low. This means the smaller the value of \(h^{2}_{PRS}\), the less difference there is between the heart attack percentages for those low polygenic risk scores groups. This explains why the PRS set with the lowest value of variance \(h^{2}_{PRS} = 0.01\) has the highest percentages of heart attack events for the first four low PRS risk groups, but the dark blue line has the lowest percentages. Similarly, with the increasing PRS variance, more and more heart attack events happen at the 80–100 percentile PRS risk group. Even though the ability of PRS to distinguish cases and controls is not that substantial under low values of \(h^{2}_{PRS}\), percentages of heart attack events still increase with increasing PRS values for both females and males. Therefore a low accuracy set of PRS is still useful risk prediction.
Proportions of the number of heart attack events in each PRS strata over the total number heart attack events from the overall dataset for a females and b males. Percentages are mean percentages calculated from 100 simulation replicates, with simulation size 500,000, and heritability as shown for each plot
The cumulative incidences of heart attack in different PRS risk groups are used to visualize the impact of PRS accuracy. Figure 9 shows the log cumulative incidence versus age for three PRS risk groups. Points used to create this figure comes from only one simulation under each value of \(h^{2}_{PRS}\). The x-axis represents age, from 0 to 69 at the last birthday. Cumulative incidence increases with age and PRS values. With increasing \(h^{2}_{PRS}\), the difference among three PRS risk groups becomes more obvious. When comparing simulated cumulative incidence with the results generated from real genotype data, the trend shown on those plots are similar to the results from Figure 2 of Maxwell et al. [41]. Their plots were created using PRS on coronary artery disease (CAD) calculated for UK Biobank participants. The UK Biobank is a large cohort study data source, including 500, 000 British participants’ genotyping data, and hundreds of variables for phenotyping information, like basic characteristics, lifestyle, medical history, and nutritional habits. Cumulative incidence numbers between their plots and Fig. 9 are quite different, as the real data set they employed has a much higher case/control ratio than our simulated data set.
3.3 Adverse selection from various purchasing scenarios
We detailed three possible adverse selection scenarios in Sect. 2.4.1. Following the assumption from Maxwell et al. [41], we also assume that HRs are used to influence simulated individuals’ probability of purchasing insurance. HRs calculated from our simulated data set are compared with the results from Maxwell et al. [41]. Following their approach, the median PRS risk group (individuals with PRS within the 40–60% percentile range), is used as the behaviour reference group in the Cox regression analysis with age as the time-dependent variable.
Table 5 lists the HRs for the highest PRS risk group (the 90–100% quantile group) under different values of \(h^{2}_{PRS}\). The second column is the average case/control ratios from 100 simulation replicates. In this study, case/control ratios are controlled by the heart attack incidence in the 4-state Markov model after a 1-year simulation. Therefore those ratios are fixed at around 0.0012 and not related to \(h^{2}_{PRS}\). The third column of this table gives mean values of HRs calculated from 100 simulation replicates and confidence intervals corresponding to 0.025 and 0.975 quantiles. HRs for the highest PRS risk group calculated from our simulated datasets are all greater than 1, and increase sharply with increasing of \(h^{2}_{PRS}\). Maxwell et al. [41] obtain the HR of CAD on UK Biobank participants for the same PRS percentile 90–100% group as 1.87, which is close to the value we find when \(h^{2}_{PRS}\) is set to 0.01. In their analysis, the case/control ratio is 1.7%, much higher than the case/control ratio in our simulation (0.12%). Because the UK Biobank participants were recruited between 2006 and 2010 and have been followed since then, the cases in their study are multi-year events. Sample size and case/control ratio both play important roles in the determination of HR size, but the study of their roles is beyond the scope of this study. Our results show that when the variance explained by PRS is high, the value of HR in the high PRS risk group will be large.
3.3.1 The increased proportion in incidence of the insured group
Under the adverse selection scenarios described, because insurance policy holders can take advantage of knowing their PRS-related results when choosing whether to purchase insurance, it is reasonable to expect that the insured group (as defined in Sect. 2.4.2) has a higher proportion of heart attack events than the overall population.
Table 6 gives the increased proportion in heart attack incidence in the insured group versus the overall simulated population for three scenarios under different values of \(h^{2}_{PRS}\). Mean increases in incidence are most obvious for scenario 2 (100% of simulated individuals know their PRS results and 1% of individuals intend to purchase insurance as a baseline measure) and less obvious for scenario 3 (10% of simulated individuals know their PRS results and 1% of individuals intend to purchase insurance as a baseline measure). Table 6 highlights the groups with confidence intervals that do not include 0, which implies that in this case there is an increase in incidence and there exists adverse selection. Those groups account for almost all three scenarios when \(h^{2}_{PRS}\) is equal to 0.3 and account for scenario 1 when \(h^{2}_{PRS}\) is equal to 0.1. Based on our results, we can say that when the accuracy of PRS is relatively low, the impact of PRS on insurers is negligible. However, as the PRS risk prediction ability increases, its impact increases sharply. In all three cases, there is an increase in the incidence of heart disease, and the severity for insurers is impacted by the proportion purchasing insurance and strongly influenced by the proportion of individuals who know their PRS results.
3.4 Premium comparison
The increased incidence in the insured group indicates that insurers will have to pay out for more heart attack events at the end of the simulated year. To balance the increased cost, insurers can increase premiums. We calculate premiums for the overall simulated population and for the insured group (the steps to identify the insured group are in Sect. 2.4.2) and then compare them. In this study, we calculate premiums for a 1-year single illness stand alone policy, which pays a lump sum of \(\pounds 10{,}000\) at the end of the policy year if the policyholder has a heart attack during the policy year. For simplicity, heart attack incidence rates from groups of interest are used to approximate the transition intensity between the healthy state and the heart attack state from Fig. 2 and then used to calculate premiums following Eq. (11). The incidence for any group of interest can be calculated from the simulated data, which is an advantage of this simulation-based study.
We first calculate premiums for each age and sex group in Table 2 without using family history as a rating factor and then calculate premiums for each age and sex group with and without family history separately.
3.4.1 Premiums for each age and sex group
Premiums for the overall simulated data set are shown in Table 7, which gives the premiums and corresponding confidence intervals for both females and males in six age groups. Numbers in this table are mean premiums and 0.025 and 0.975 quantile values from 100 simulation replicates, each with sample size 500, 000 and heritability 0.1. Age and sex-based group incidence is not affected by the size of \(h^{2}_{PRS}\), so premiums calculated using alternative values of \(h^{2}_{PRS}\) are very similar to each other. Heart attack incidences for females and males used to calculate premiums in Table 7 are in Table 8 for reference. The youngest age group is removed because the female group has heart attack incidence of zero in simulations. Males have higher incidence, so their premiums are higher than females at all ages.
We defined three adverse selection scenarios where insurance applicants took advantage of knowing their PRS results when purchasing insurance. Incidence rates in the insured group are higher than the overall simulated data (Table 6). Premiums calculated for the insured group are shown in Table 9, which contains premiums calculated for all three scenarios under each value of \(h^{2}_{PRS}\). The mean values in bold emphasize the groups where the mean premium for the insured group is within the premium confidence interval calculated for the overall simulated data set in Table 7. For those groups, this may suggest that there is a reduced need to increase premiums to offset the risk of adverse selection.
This outcome occurs for almost all groups when \(h^{2}_{PRS}\) is equal to 0.01 and the majority groups from Scenario 3 when \(h^{2}_{PRS}\) is not equal to 0.3. Within this table, comparing Scenarios 1 and 2, their values are similar, but Scenario 2 has slightly higher values. Two possible reasons can explain this. Firstly, given that Scenario 1 has 10% baseline purchasing insurance while Scenario 2 only has 1%, we can say the severity of adverse selection due to PRS can be offset to some extent by a larger insurance pool. Secondly, when the HR-based proportion purchasing insurance is calculated as being greater than \(100\%\), we use \(100\%\) (Sect. 2.4.2). However, this may disguise the true severity of adverse selection in Scenario 1 because when HR is greater than 10, an insurance applicant is assumed to buy only one insurance policy with probability 100%. We do not consider the scenarios where insurance applicants buy multiple policies.
The proportion of increased premiums were plotted in Fig. 10. Here, the − 100% change refers to the situation where there are no heart attack events simulated in the insured group. The premium increase proportions are smaller in males than in females when \(h^{2}_{PRS}\) is equal to 0.1 or 0.3. This can be explained by the higher base line premiums for males than females, which is caused by the higher heart attack incidence rates in males than females. Based on this figure, we can say that premiums for female insurance applicants should be increased more than those for male insurance applicants to offset the adverse selection brought about by knowledge of PRS.
3.4.2 Premiums for each age and sex group with and without family history
Table 10 contains premiums calculated using incidence rates of the overall simulated data for each age and sex group with and without family history respectively. We then calculated premiums for the insured groups for three adverse selection scenarios under each value of \(h^{2}_{PRS}\). Figure 11 shows the proportion of premium increases compared with premiums in Table 10. Solid and dashed lines are used for groups with family history and without family history respectively. Like Fig. 10, the -100% change refers to the situation where there are no heart attack events simulated in the insured group. Also, the adverse selection caused by knowing PRS results is negligible when the accuracy of PRS is very low (\(h^{2}_{PRS}\) = 0.01). Additionally, premiums for female insurance applicants should be increased more than those for male insurance applicants to offset the adverse selection brought about by policyholders knowing PRS.
Regarding family history, Fig. 11 tells us that premiums should be increased further for groups without family history than groups with family history, to offset the impact of PRS. This is because the number of heart attack events is higher in the group with family history than in the group without family history, so the premium is already higher in the group with family history than in the group without a family history. Family history contains some degree of genetic information, so pricing insurance products that use family history as an indicator of risk already offsets genetic risk to some extent.
4 Conclusion
With the ongoing development of genetic research our understanding of the genetic architectures of diseases will be enhanced, as well as the ability to predict disease risk. The accompanying progress of direct-to-customer testing will make it easier for the general public to understand or even to predict their own risk of diseases. PRS is one such genetic test result that can add to people’s information on their disease risk. PRS has the ability to identify high-risk groups when used independently or jointly with other risk factors and the potential to inform early interventions, which may change future morbidity and mortality. Although PRS is not very accurate in predicting risk at the individual level at present, the impact of PRS on the foundations of the insurance industry will become increasingly evident as the technology evolves.
Employing simulated data, this study introduces individual-level PRS into the actuarial analysis framework using heart attack as a case study. Parent and offspring disease liabilities are simulated following a multivariate normal distribution, with the real-world values of heritability and the relationship of parents and their offspring determining the elements in the multivariate covariance matrix. Disease states for both parents and offspring are determined using the liability threshold model, which links disease prevalence and a continuous disease liability. The age and sex specific disease prevalence is calculated from the transition intensities under a 4-state heart attack Markov model. We validated our simulated datasets and confirmed that our simulation mimicked the real world scenarios well. Therefore, this simulation-based model provides a framework for insurers to measure the impact of PRS on life and health insurance in various scenarios.
Using the simulated data, insurers can explore any scenario of interest and quantify the corresponding impact on their business. We explored three possible adverse selection scenarios, where insurance policy holders took advantage of knowing their PRS results (HR values in this study) when purchasing insurance. Then we measured the increased incidence in the insured group versus the overall simulated population for each scenario of interest. As a response, insurers could increase premiums to balance the increased heart attack events among the insured population. We calculated the extent of increased premiums after adverse selection happens. The accuracy of PRS has the most significant impact on insurers and the proportion of individuals knowing their PRS also impacts the extent of the increased premiums. Comparing the increase in premiums between with and without family history groups, we found that premiums should be increased further for groups without family history to offset the impact of PRS.
There are limitations of this study. The first limitation is that our current model only works on homogeneous populations and only applies to one disease. Narrowing the participants in statistical genetics studies to samples with a single ancestry aids the accuracy in the research findings [57]. European ancestry samples are the most common participants in medical genetics research, including the calculation of PRS, while European ancestry-derived polygenic scores have lower predictive performance in samples of non-European ancestry [8]. However, insurance policyholders come from diverse populations. Duncan et al. [8] has called for carrying out large-scale genetic association studies in diverse human populations. Similarly, insurance policies usually cover multiple diseases rather than a single disease. A model including PRS from multiple diseases should be designed when it comes to measuring the impact of PRS on a specific type of insurance product.
Second, this study examined the impact of PRS from only one type of risk indicator - hazard ratios - but there are other types of risk indicators available to DTC test clients. Clients from the same insurance market may receive different indicators to inform them of their disease risk, including odds ratios, relative risks, etc. With more and more customers possessing details on their genetic profiles, follow-up studies like [64] are needed. From a follow- up study tracking individuals participating in a clinical trial of genetic testing for Alzheimer’s disease (AD), Zick et al. [64] found evidence that genetic testing results can alter people’s insurance purchasing decisions. Participants who tested positive were 5.76 times more likely to change their long-term care insurance than individuals who did not receive the AD related risk gene disclosure. Understanding how people react to their PRS results can help insurers more accurately measure the impact of PRS.
Finally, we assume that the disease liability of parents and offspring remains the same through their lifetime. Disease liability combines the genetic and non-genetic components, including the interaction between them. Even if we assume that the human genetic profile is fixed throughout life, disease liability is susceptible to change due to factors such as the environment and lifestyle. Additionally, Jiang et al. [25] found evidence for age-varying relative risk profiles in common diseases, which means genetic risk factors have stronger influence on younger populations compared to older ones. In this study, 1 year is chosen as the simulation period because it is a reliable assumption that disease liability stays approximately constant within 1 year. If we want to expand the simulation period to multiple years, the change in disease liability with time should be considered as well as evolution in family history.
References
Bachmann JM, Willis BL, Ayers CR, Khera A, Berry JD (2012) Association between family history and coronary heart disease death across long-term follow-up in men: the cooper center longitudinal study. Circulation 125(25):3092–3098
Bélisle-Pipon JC, Vayena E, Green RC, Cohen IG (2019) Genetic testing, insurance discrimination and medical research: what the united states can learn from peer countries. Nat Med 25(8):1198–1204. https://doi.org/10.1038/s41591-019-0534-z
Brown TA (2018) Genomes 4. Garland science, New York. https://doi.org/10.1201/9781315226828
Chatterjee I, Macdonald AS, Tapadar P, Thomas RG (2021) When is utilitarian welfare higher under insurance risk pooling? Insur Math Econ 101:289–301
Choi SW, Mak TSH, O’Reilly P (2020) Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc 1:1–14. https://doi.org/10.1038/s41596-020-0353-1
Claussnitzer M, Cho JH, Collins R, Cox NJ, Dermitzakis ET, Hurles ME, Kathiresan S, Kenny EE, Lindgren CM, MacArthur DG et al (2020) A brief history of human disease genetics. Nature 577(7789):179–189. https://doi.org/10.1038/s41586-019-1879-7
Concordat: Government of the United Kingdom and Association of British Insurers: Concordat and Moratorium on Genetics and Insurance (2014). https://www.abi.org.uk/globalassets/sitecore/files/documents/publications/public/2014/genetics/concordat-and-moratorium-on-genetics-and-insurance.pdf
Duncan L, Shen H, Gelaye B, Meijsen J, Ressler K, Feldman M, Peterson R, Domingue B (2019) Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun 10(1):1–9. https://doi.org/10.1038/s41467-019-11112-0
Euesden J, Lewis CM, O’Reilly PF (2014) PRSice: polygenic risk score software. Bioinformatics 31(9):1466–1468. https://doi.org/10.1093/bioinformatics/btu848
Falconer DS (1965) The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann Hum Genet 29(1):51–76
Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics. Longman, Prentice Hall, Essex
Folkersen L, Pain O, Ingasson A, Werge T, Lewis CM, Austin J (2019) Impute me: an open source, non-profit tool for using data from DTC genetic testing to calculate and interpret polygenic risk scores. BioRxiv 11:861831. https://doi.org/10.3389/fgene.2020.00578
Francis LP (2010) You are born with your genes: justice and protection against discrimination in the use of genetic information. Mt Sinai J Med 77(2):188–196. https://doi.org/10.1002/msj.20170
Ge T, Chen CY, Ni Y, Feng Y-CA, Smoller JW (2019) Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun 10(1):1776. https://doi.org/10.1038/s41467-019-09718-5
GenoPred: Converting polygenic score to absolute scale (2022). https://opain.github.io/GenoPred/PRS_to_Abs_tool.html
Gutiérrez C, Macdonald AS (2003) Adult polycystic kidney disease and critical illness insurance. North Am Actuar J 7(2):93–115. https://doi.org/10.1080/10920277.2003.10596092
Gutiérrez C, Macdonald AS (2007) Adult polycystic kidney disease and insurance: a case study in genetic heterogeneity. North Am Actuar J 11(1):90–118. https://doi.org/10.1080/10920277.2007.10597439
Hinds DA, Stokowski RP, Patil N, Konvicka K, Kershenobich D, Cox DR, Ballinger DG (2004) Matching strategies for genetic association studies in structured populations. Am J Hum Genet 74(2):317–325. https://doi.org/10.1086/381716
Hock Gui E, Lu B, Macdonald A, Waters H (2006) Wekwete C (2006) The genetics of breast and ovarian cancer III: a new model of family history with insurance applications. Scand Actuar J 6:338–367. https://doi.org/10.1080/03461230601026635
Howard R (2014) Genetic testing model: if underwriters had no access to known results. Report to Canadian Institute of Actuaries Research Committee, July 2014
Howard R (2016) Genetic testing model for CI: if underwriters of individual critical illness insurance had no access to known results of genetic tests. Report to Canadian Institute of Actuaries Research Committee, January 2016
Huang J, Ling CX (2005) Using AUC and accuracy in evaluating learning algorithms. IEEE Trans Knowl Data Eng 17(3):299–310
Hujoel ML, Gazal S, Loh P-R, Patterson N, Price AL (2020) Liability threshold modeling of case-control status and family history of disease increases association power. Nat Genet 52(5):541–547. https://doi.org/10.1038/s41588-020-0613-6
Human Genome Project Information Archive: about the human genome project (1990). https://web.ornl.gov/sci/techresources/Human_Genome/project/index.shtml
Jiang X, Holmes C, McVean G (2021) The impact of age on genetic risk for common diseases. PLoS Genet 17(8):1009723. https://doi.org/10.1371/journal.pgen.1009723
Joly Y, Burton H, Knoppers BM, Feze IN, Dent T, Pashayan N, Chowdhury S, Foulkes W, Hall A, Hamet P et al (2014) Life insurance: genomic stratification and risk classification. Eur J Hum Genet 22(5):575–579. https://doi.org/10.1038/ejhg.2013.228
Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, Chasman DI, Baber U, Mehran R, Rader DJ et al (2016) Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med 375(24):2349–2358. https://doi.org/10.1056/NEJMoa1605086
Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT et al (2018) Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 50(9):1219–1224. https://doi.org/10.1038/s41588-018-0183-z
Lee SH, Wray NR, Goddard ME, Visscher PM (2011) Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet 88(3):294–305. https://doi.org/10.1016/j.ajhg.2011.02.002
Lewis CM, Vassos E (2020) Polygenic risk scores: from research tools to clinical instruments. Genome Med 12:1–11. https://doi.org/10.1186/s13073-020-00742-5
Lewis AC, Green RC (2021) Polygenic risk scores in the clinic: new perspectives needed on familiar ethical issues. Genome Med 13(1):1–10. https://doi.org/10.1186/s13073-021-00829-7
Lloyd-Jones DM, Nam B-H, D’Agostino RB Sr, Levy D, Murabito JM, Wang TJ, Wilson PW, O’Donnell CJ (2004) Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA 291(18):2204–2211
Macdonald AS, Waters HR (2003) Wekwete CT (2003) The genetics of breast and ovarian cancer I: a model of family history. Scand Actuar J 1:1–27
Macdonald AS, Waters HR (2003) Wekwete CT (2003) The genetics of breast and ovarian cancer II: a model of critical illness insurance. Scand Actuar J 1:28–50
Macdonald AS (2004) Huntington’s disease, critical illness insurance and life insurance. Scand Actuar J 2004(4):279–313. https://doi.org/10.1080/03461230310016992
Macdonald A, Pritchard D, Tapadar P (2006) The impact of multifactorial genetic disorders on critical illness insurance: a simulation study based on UK biobank. ASTIN Bull J IAA 36(2):311–346. https://doi.org/10.1017/S0515036100014537
Macdonald A, Tapadar P (2010) Multifactorial genetic disorders and adverse selection: epidemiology meets economics. J Risk Insur 77(1):155–182. https://doi.org/10.1111/j.1539-6975.2009.01342.x
Mak TSH, Porsch RM, Choi SW, Zhou X, Sham PC (2017) Polygenic scores via penalized regression on summary statistics. Genet Epidemiol 41(6):469–480. https://doi.org/10.1002/gepi.22050
Mars N, Lindbohm JV, della Briotta Parolo P, Widén E, Kaprio J, Palotie A, Ripatti S et al (2022) Systematic comparison of family history and polygenic risk across 24 common diseases. Am J Hum Genet
Martin AR, Daly MJ, Robinson EB, Hyman SE, Neale BM (2019) Predicting polygenic risk of psychiatric disorders. Biol Psychiatry 86(2):97–109. https://doi.org/10.1016/j.biopsych.2018.12.015
Maxwell JM, Russell RA, Wu HM, Sharapova N, Banthorpe P, O’Reilly PF, Lewis CM (2021) Multifactorial disorders and polygenic risk scores: predicting common diseases and the possibility of adverse selection in life and protection insurance. Ann Actuar Sci 15(3):488–503. https://doi.org/10.1017/S1748499520000226
McPherson R, Tybjaerg-Hansen A (2016) Genetics of coronary artery disease. Circ Res 118(4):564–578. https://doi.org/10.1161/circresaha.115.306566
McCormick A (1995) Morbidity statistics from general practice. Fourth national study 1991–1992. Office of population censuses and surveys
Meisner A, Chatterjee N (2019) Disease risk models. In: Balding DJ, Moltke I, Marioni J (eds) Handbook of statistical genomics. Wiley, New York, pp 815–841
Pain O, Gillett AC, Austin JC, Folkersen L, Lewis CM (2022) A tool for translating polygenic scores onto the absolute scale using summary statistics. Eur J Hum Genet. https://doi.org/10.1038/s41431-021-01028-z
Plomin R, Von Stumm S (2021) Polygenic scores: prediction versus explanation. Mol Psychiatry. https://doi.org/10.1038/s41380-021-01348-y
Privé F, Arbel J, Vilhjálmsson BJ (2020) Ldpred2: better, faster, stronger. Bioinformatics 36(22–23):5424–5431. https://doi.org/10.1093/bioinformatics/btaa1029
Population Pyramid (2022) Population pyramids of the world from 1950 to 2100. https://opain.github.io/GenoPred/PRS_to_Abs_tool.html
Prince AE (2018) Political economy, stakeholder voices, and saliency: lessons from international policies regulating insurer use of genetic information. J Law Biosci 5(3):461–494
Purcell S, Wray N, Stone J, Visscher P, O’Donovan M, Sullivan P et al (2009) Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature [Internet] 460(7256):748–52
Roy R, Chun J, Powell SN (2012) Brca1 and brca2: different roles in a common pathway of genome protection. Nat Rev Cancer 12(1):68–78. https://doi.org/10.1038/nrc3181
So H-C, Kwan JS, Cherny SS, Sham PC (2011) Risk prediction of complex diseases from family history and known susceptibility loci, with applications for cancer screening. Am J Hum Genet 88(5):548–565. https://doi.org/10.1016/j.ajhg.2011.04.001
So H-C, Sham PC (2017) Improving polygenic risk prediction from summary statistics by an empirical Bayes approach. Sci Rep 7:41262. https://doi.org/10.1038/srep41262
Swiss Re Institute (2019) Genetic testing adverse selection. https://www.swissre.com/institute/research/sonar/sonar2019/sonar2019-genetic-testing-adverse-selection.html
Tapadar P (2007) The impact of multifactorial genetic disorders on long-term insurance. PhD thesis, Heriot-Watt University
Tenesa A, Haley CS (2013) The heritability of human disease: estimation, uses and abuses. Nat Rev Genet 14(2):139–149. https://doi.org/10.1038/nrg3377
Tian C, Gregersen PK, Seldin MF (2008) Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet 17(R2):143–150. https://doi.org/10.1093/hmg/ddn268
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J (2017) 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet 101(1):5–22. https://doi.org/10.1016/j.ajhg.2017.06.005
Vilhjálmsson BJ, Yang J, Finucane HK, Gusev A, Lindström S, Ripke S, Genovese G, Loh P-R, Bhatia G, Do R et al (2015) Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet 97(4):576–592. https://doi.org/10.1016/j.ajhg.2015.09.001
Vukcevic D, Chen J (2017) Thinking about life insurance through a genetic lens. Actuaries Summit, Melbourne
Why fatal heart disease is striking middle-aged patients younger and more often. https://utswmed.org/medblog/why-fatal-heart-disease-striking-middle-aged-patients-younger-and-more-often/. Accessed 01 Dec 2022
Widén E, Junna N, Ruotsalainen S, Surakka I, Mars N, Ripatti P, Partanen JJ, Aro J, Mustonen P, Tuomi T et al (2022) How communicating polygenic and clinical risk for atherosclerotic cardiovascular disease impacts health behavior: an observational follow-up study. Circ Genom Precis Med. https://doi.org/10.1161/CIRCGEN.121.003459
Wray NR, Lin T, Austin J, McGrath JJ, Hickie IB, Murray GK, Visscher PM (2021) From basic science to clinical application of polygenic risk scores: a primer. JAMA Psychiatry 78(1):101–109. https://doi.org/10.1001/jamapsychiatry.2020.3049
Zick CD, Mathews CJ, Roberts JS, Cook-Deegan R, Pokorski RJ, Green RC (2005) Genetic testing for Alzheimer’s disease and its impact on insurance purchasing behavior. Health Affairs 24(2):483–490. https://doi.org/10.1377/hlthaff.24.2.483
Acknowledgements
This publication has emanated from research conducted with funding from the Science Foundation Ireland under Grant number [SFI/12/RC/2289_P2] and with financial support from the Society of Actuaries in Ireland. For the purpose of Open Access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission.
Funding
Open Access funding provided by the IReL Consortium
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No potential competing interest was reported by the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhao, J., Salter-Townshend, M. & O’Hagan, A. A simulation study for multifactorial genetic disorders to quantify the impact of polygenic risk scores on critical illness insurance. Eur. Actuar. J. 13, 775–813 (2023). https://doi.org/10.1007/s13385-023-00345-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13385-023-00345-5